|
Putative alpha helix
|
Physico-chemical properties
|
Plots from HeliQuest
|
|
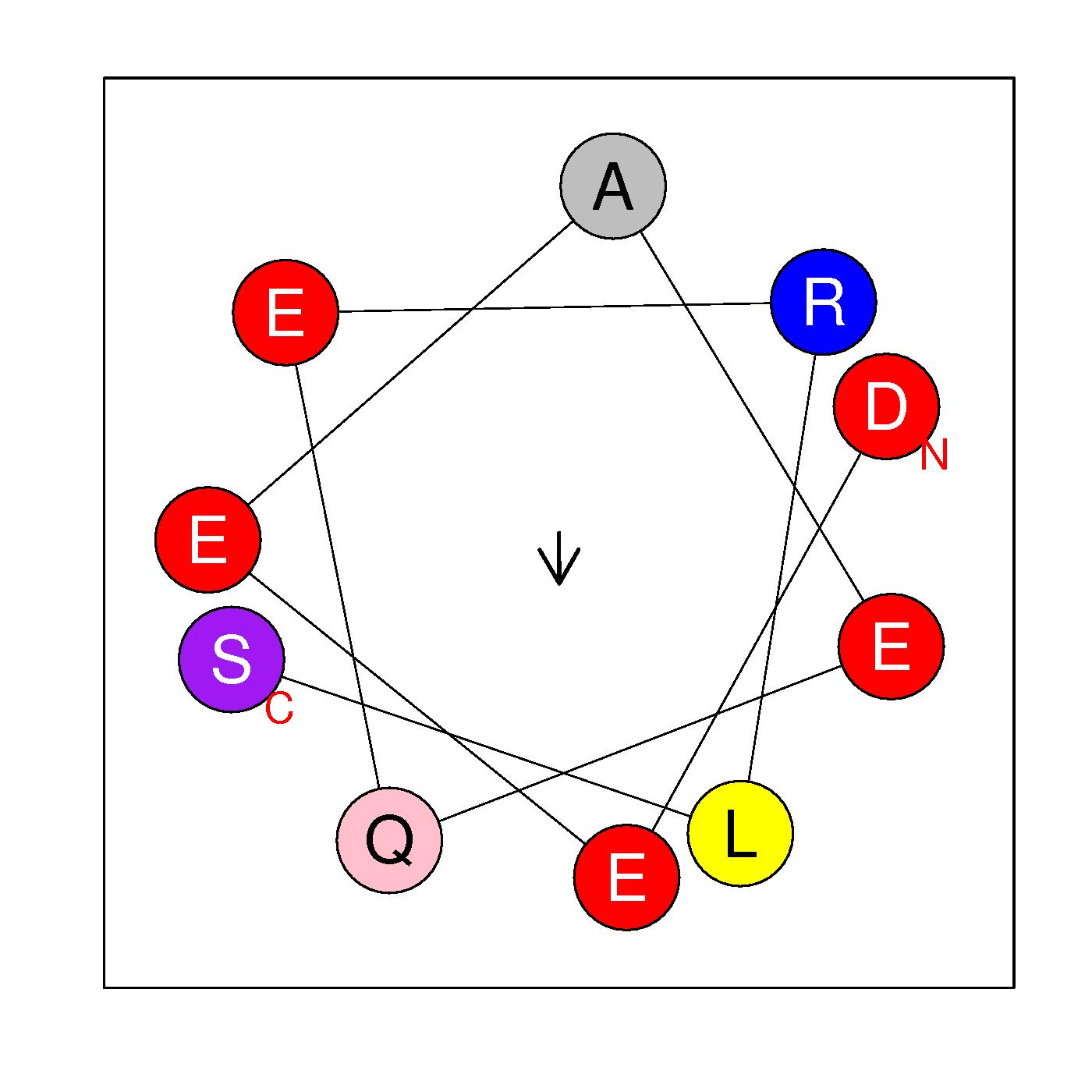
α0
DEEAEQERLS
(3-12)
|
H=-0.259 μH=0.144
Z=-4
|
|
|
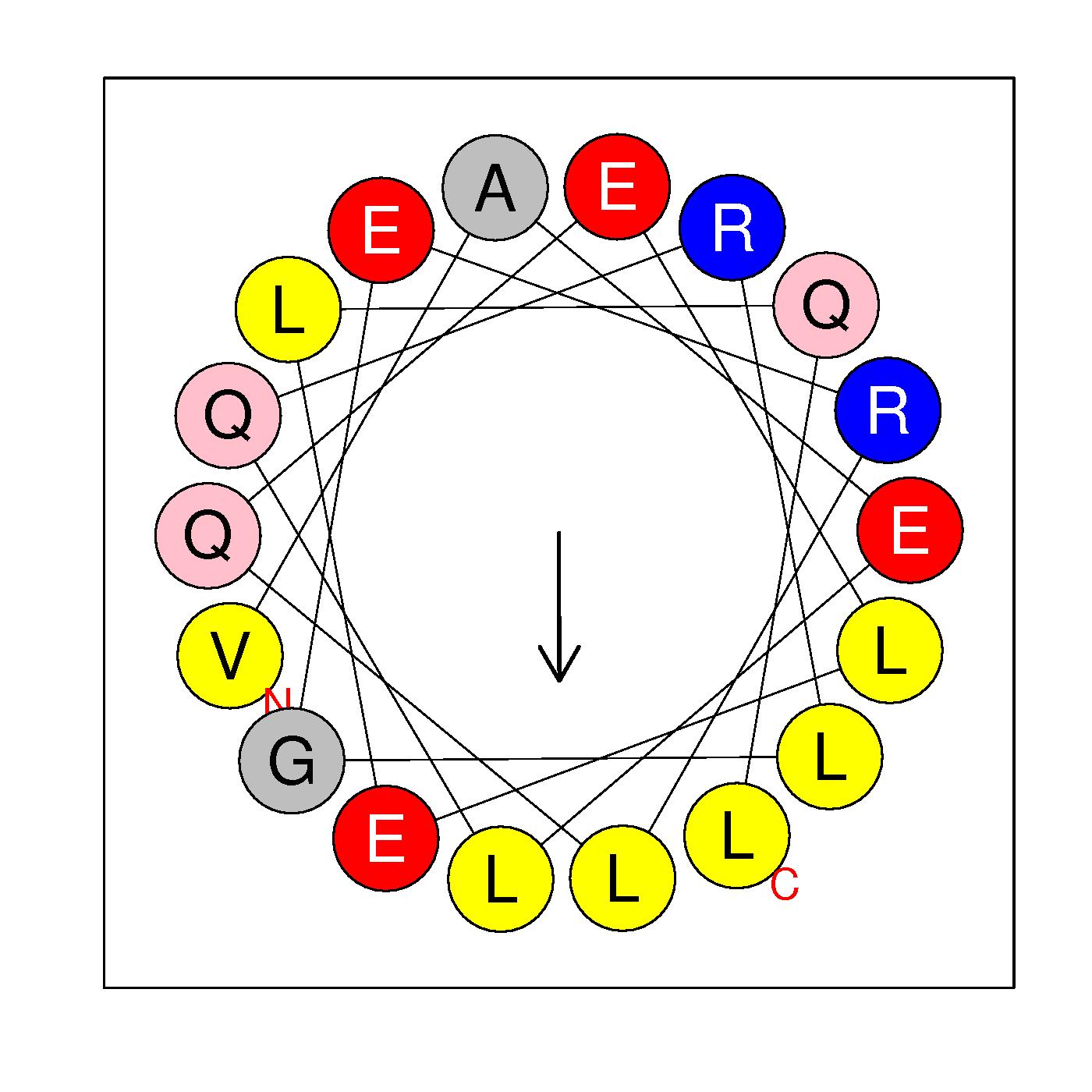
α1
VAELQRLGERLQELELQL
(19-36)
Very leucine zipper like
|
H=0.361 μH=0.421
Z=-2
Hydrophobic face L L L L L
|
|
|
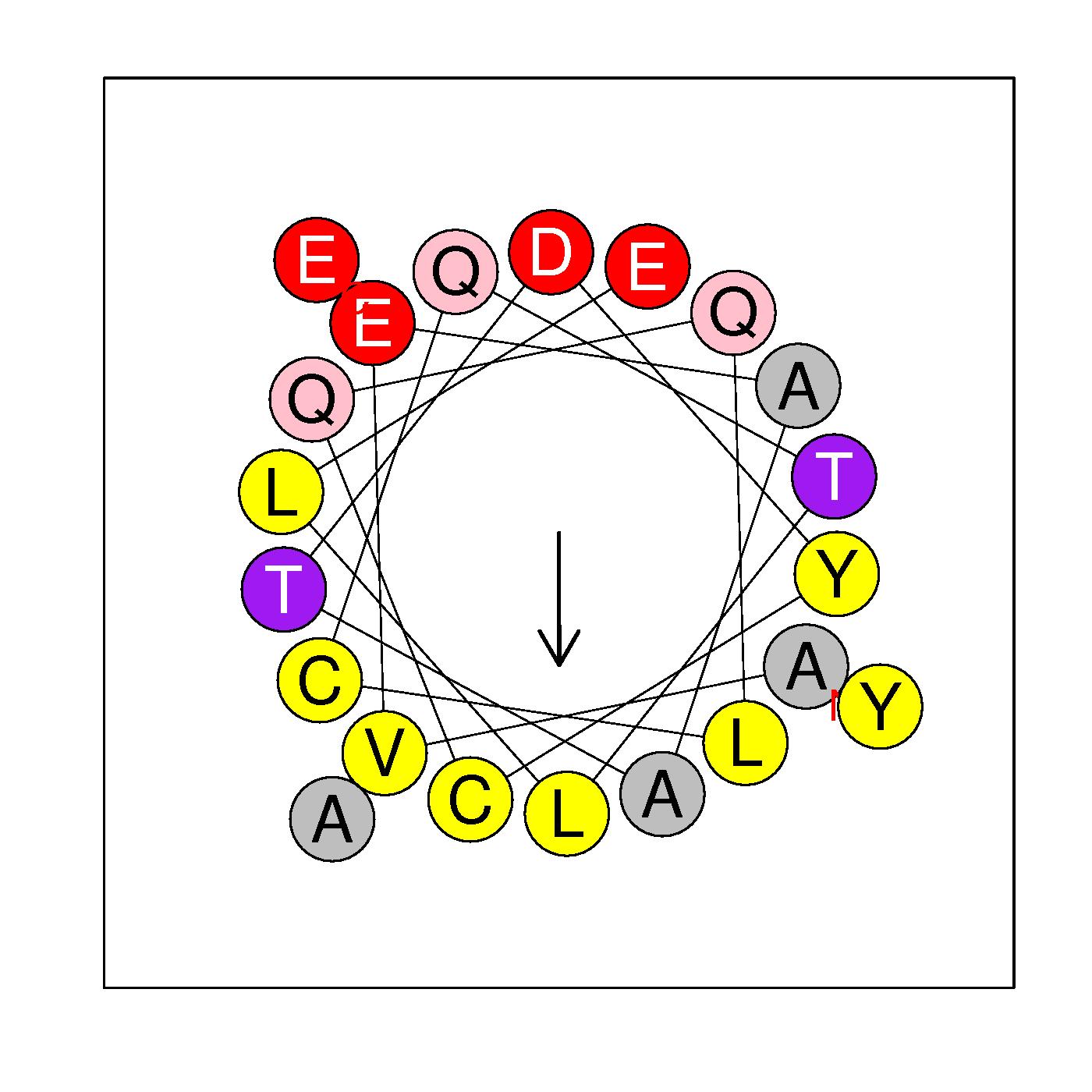
α2
AVEAATDYCQQLCQTLLEYAE
(43-63)
|
H=0.463 μH=0.470
Z=-4
Hydrophobic face Y A Y L A L
|
|
|
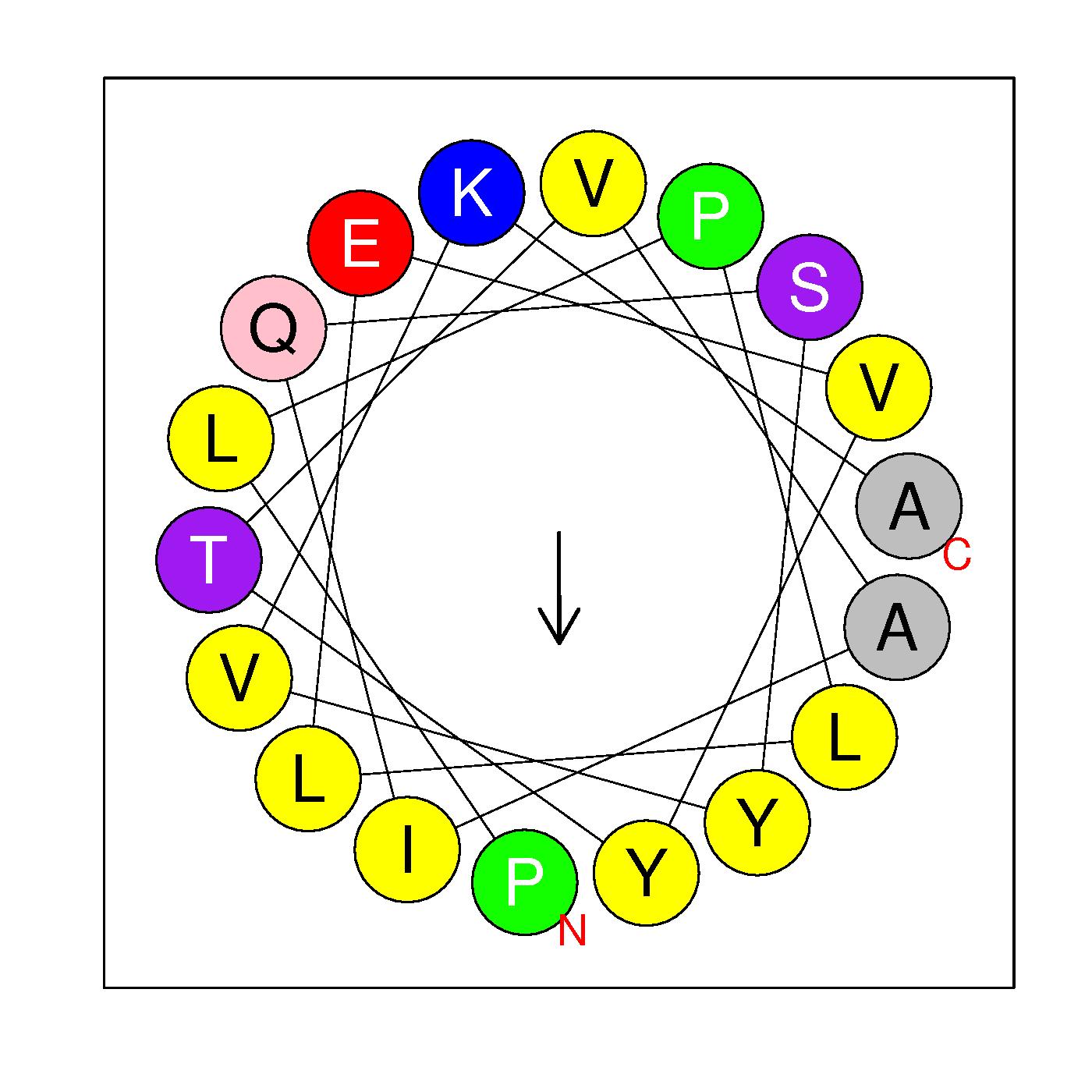
α3
PLPLLEVYTVAIQSYVKA
(71-88)
|
H=0.717 μH=0.313
Z=0
Hydrophobic face V A A L Y Y P I L V
|
|
|
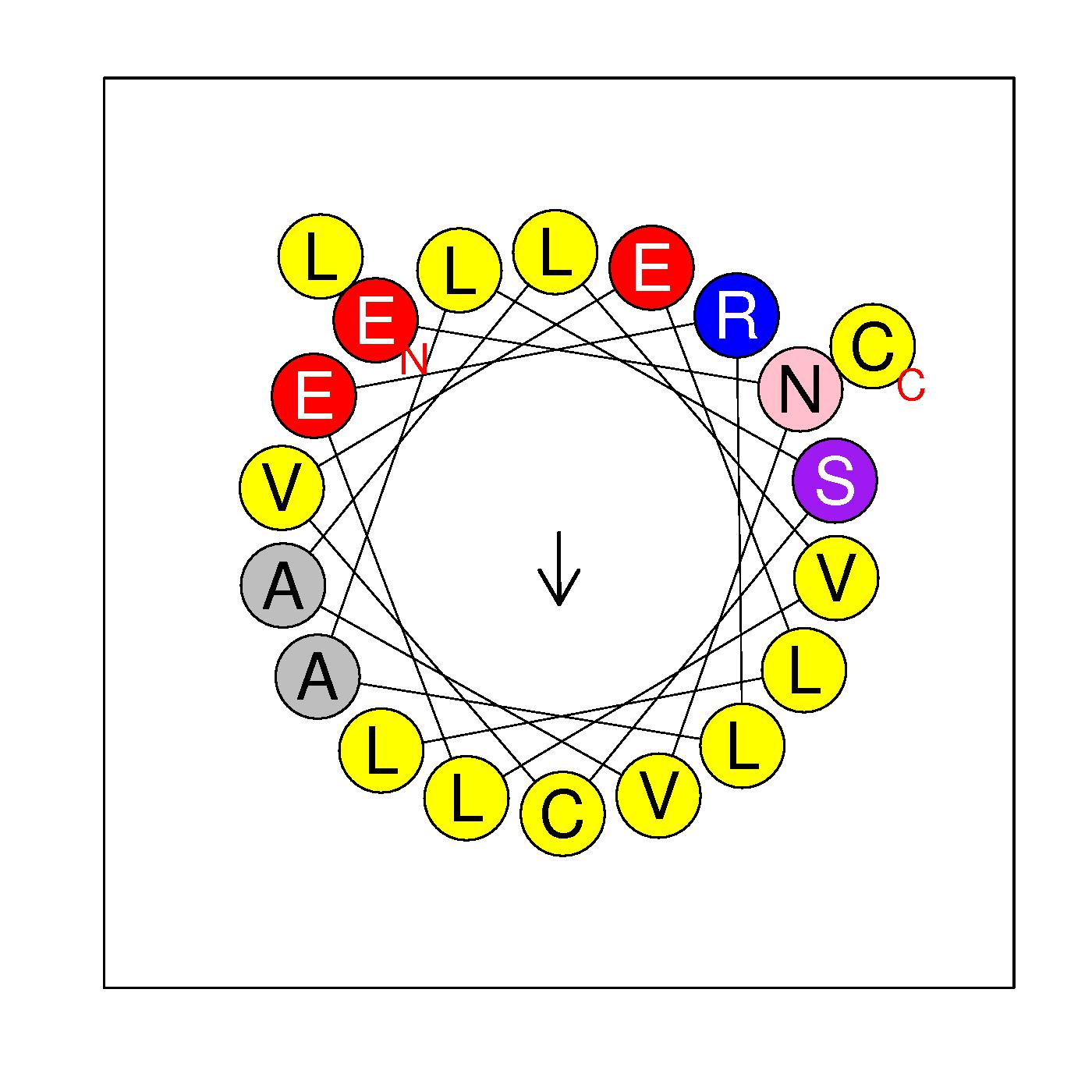
α4
ENVALVLERLALSCVELLLC
(97-116)
|
H=0.784 μH=0.254
Z=-2
Hydrophobic face L L A A V
|
|
|
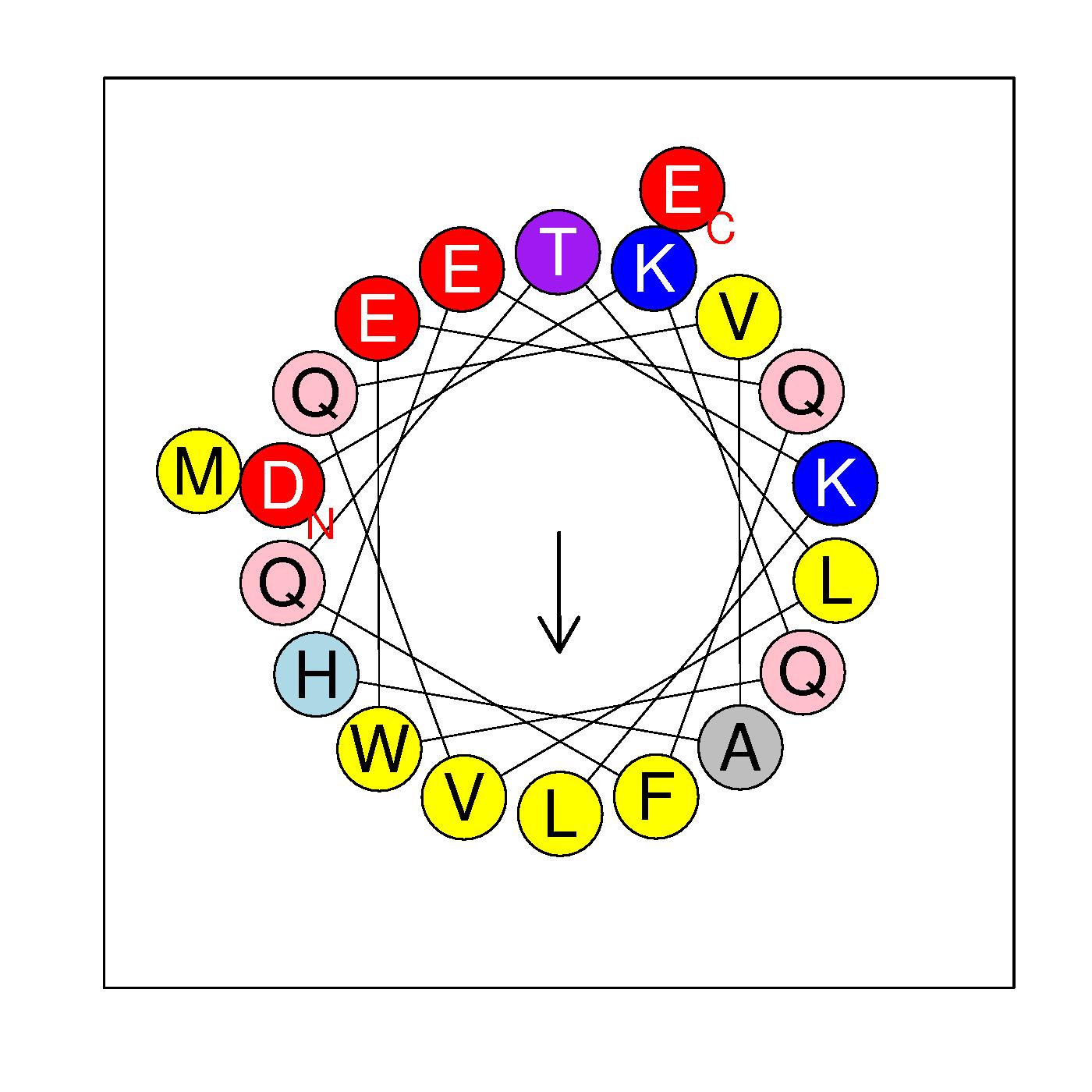
α5
DKQWEQFQTLVQVAHEKLME
(123-142)
|
H=0.313 μH=0.422
Z=-2
Hydrophobic face A F L V W
|
|
|
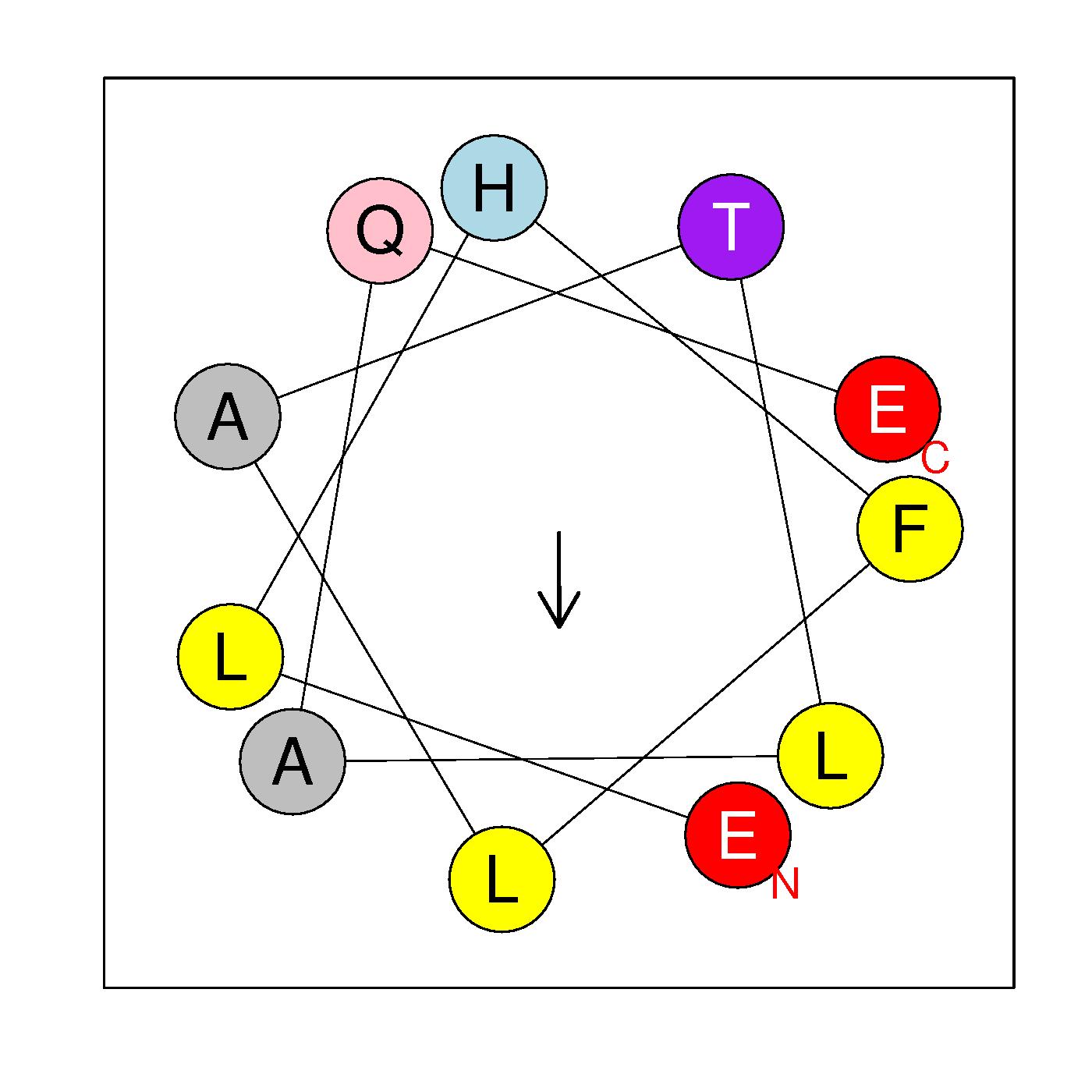
α6
ELHFLATLAQE
(147-157)
|
H=0.582 μH=0.268
Z=-2
|
|
|
α7
PVLCTIL
(164-170)
|
H=1.277
Z=0
|
|
|
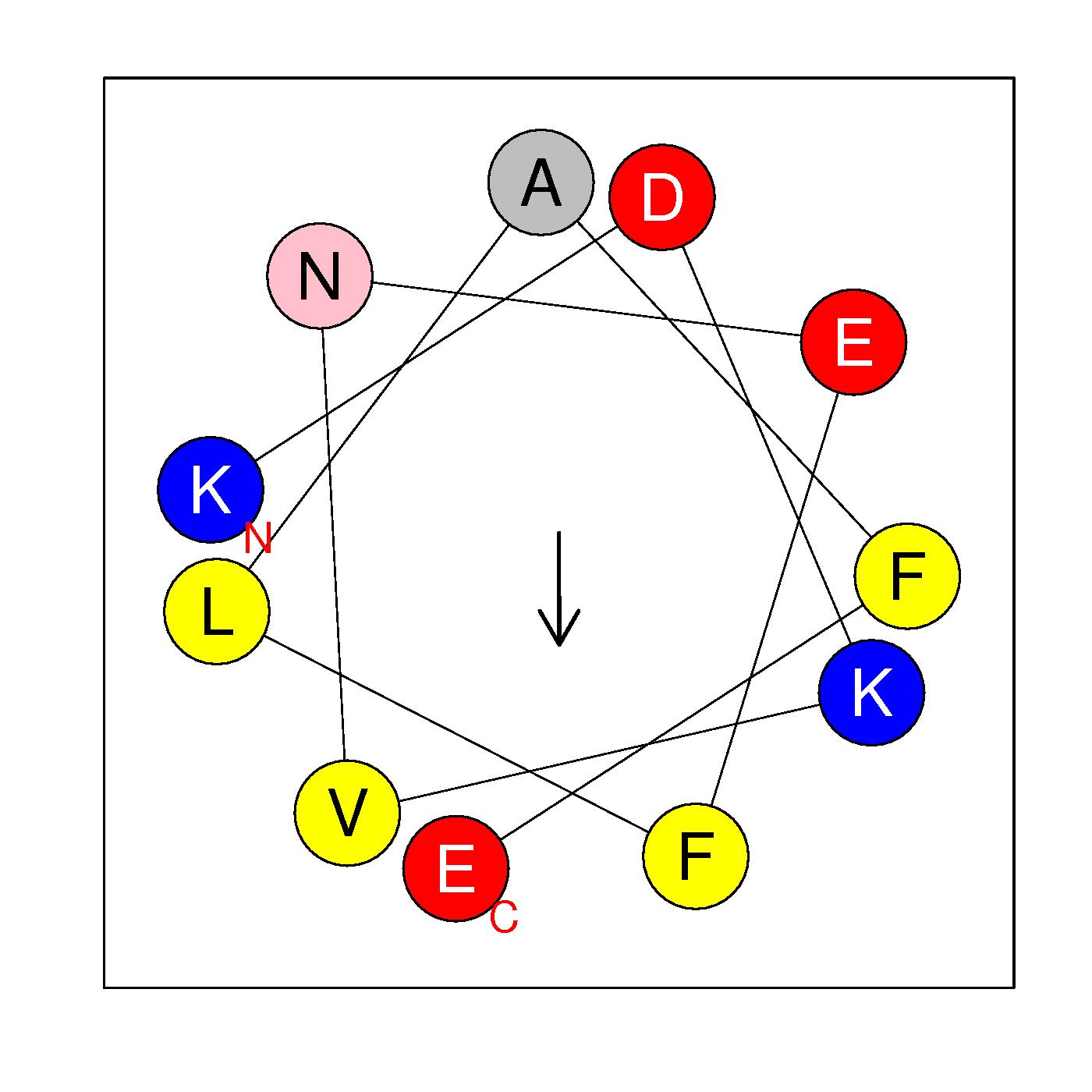
α8
KDKVNEFLAFE
(177-187)
|
H=0.198 μH=0.318
Z=-1
|
|
|
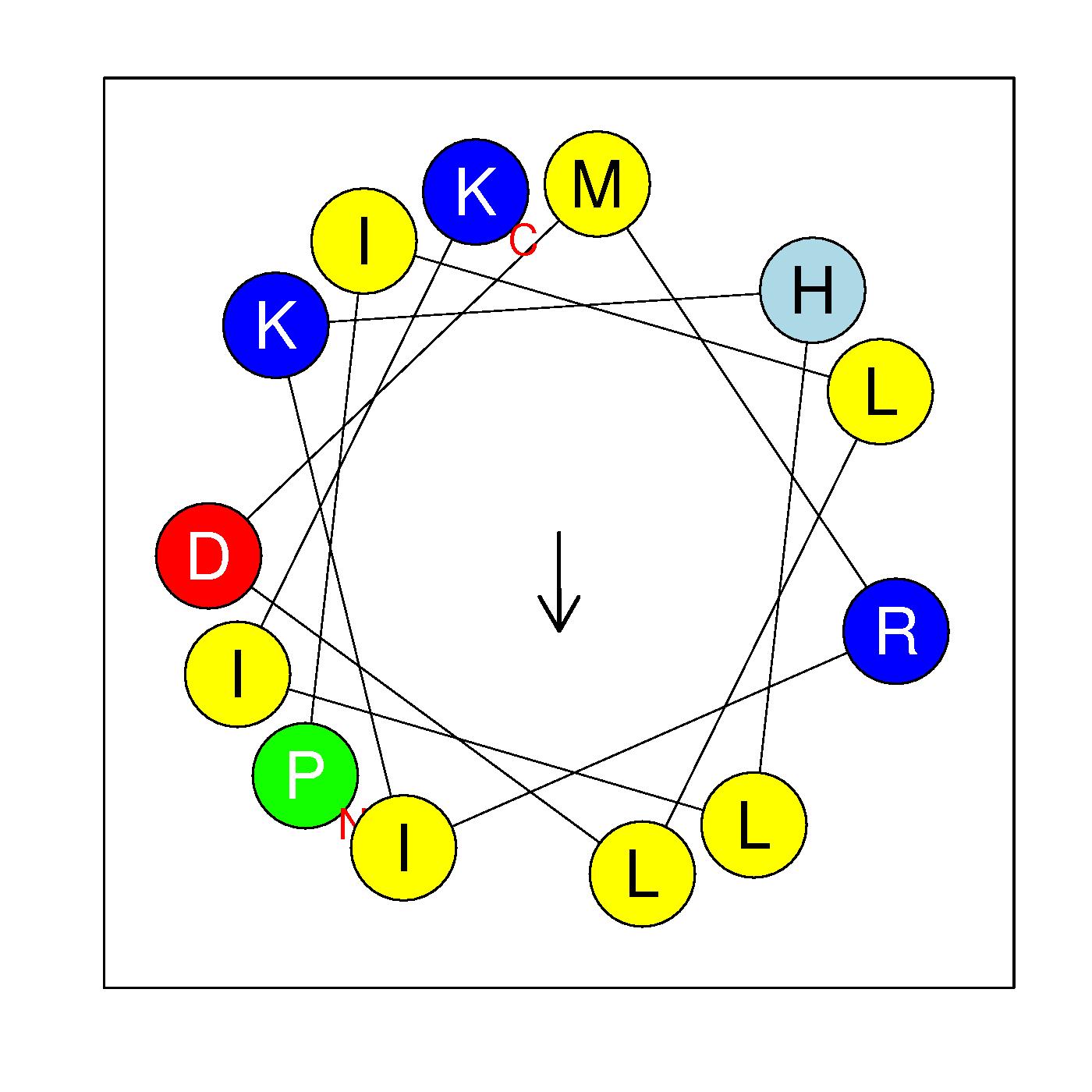
α9
PILLDMRIKHLIK
(189-201)
|
H=0.678 μH=0.279
Z=2
|
|
|
α10
LSQATALAKLCS
(205-216)
|
H=0.545 μH=0.329
Z=1
|
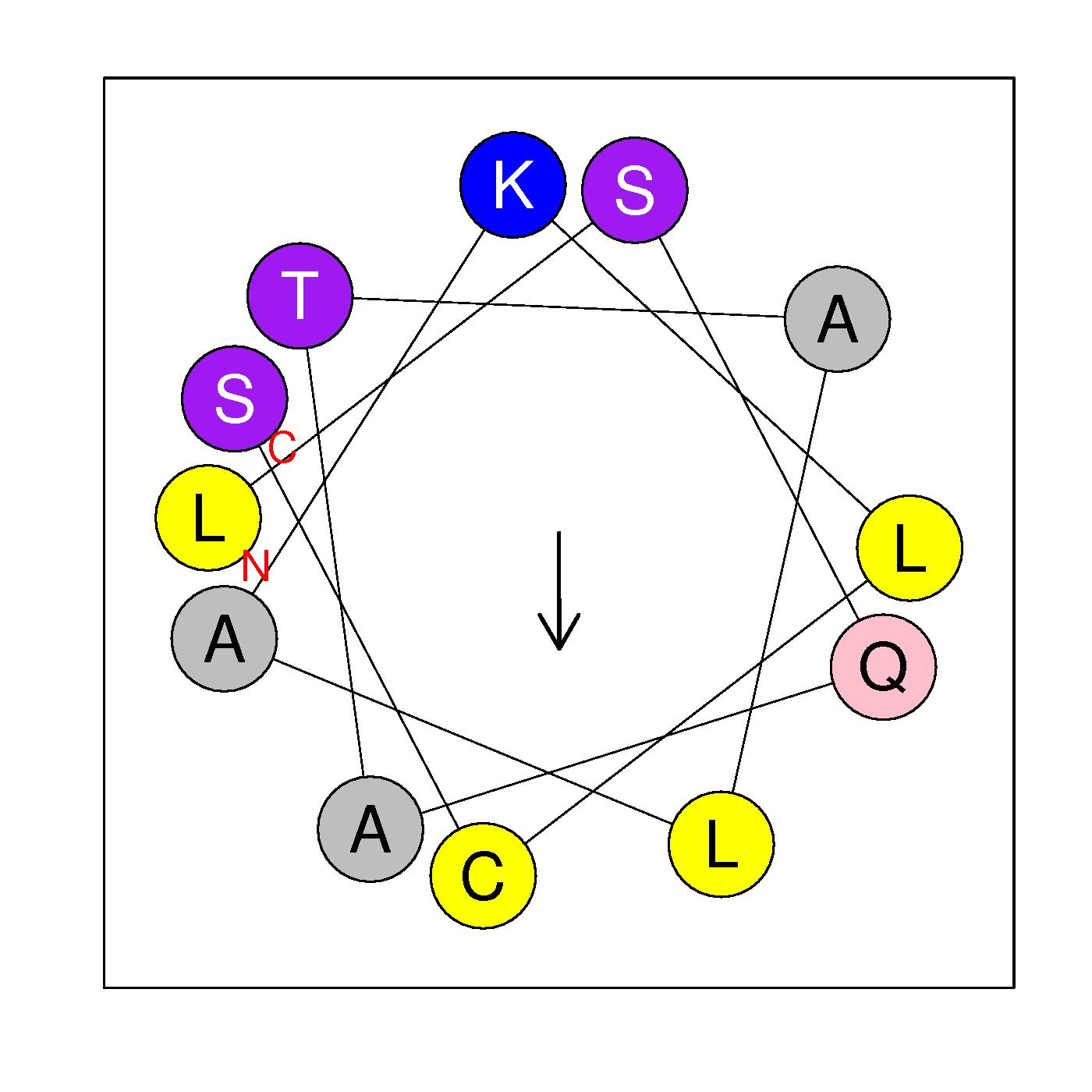
|
|
α11
FKQTYLVCLCT
(227-237)
|
H=0.887 μH=0.130
Z=1
|
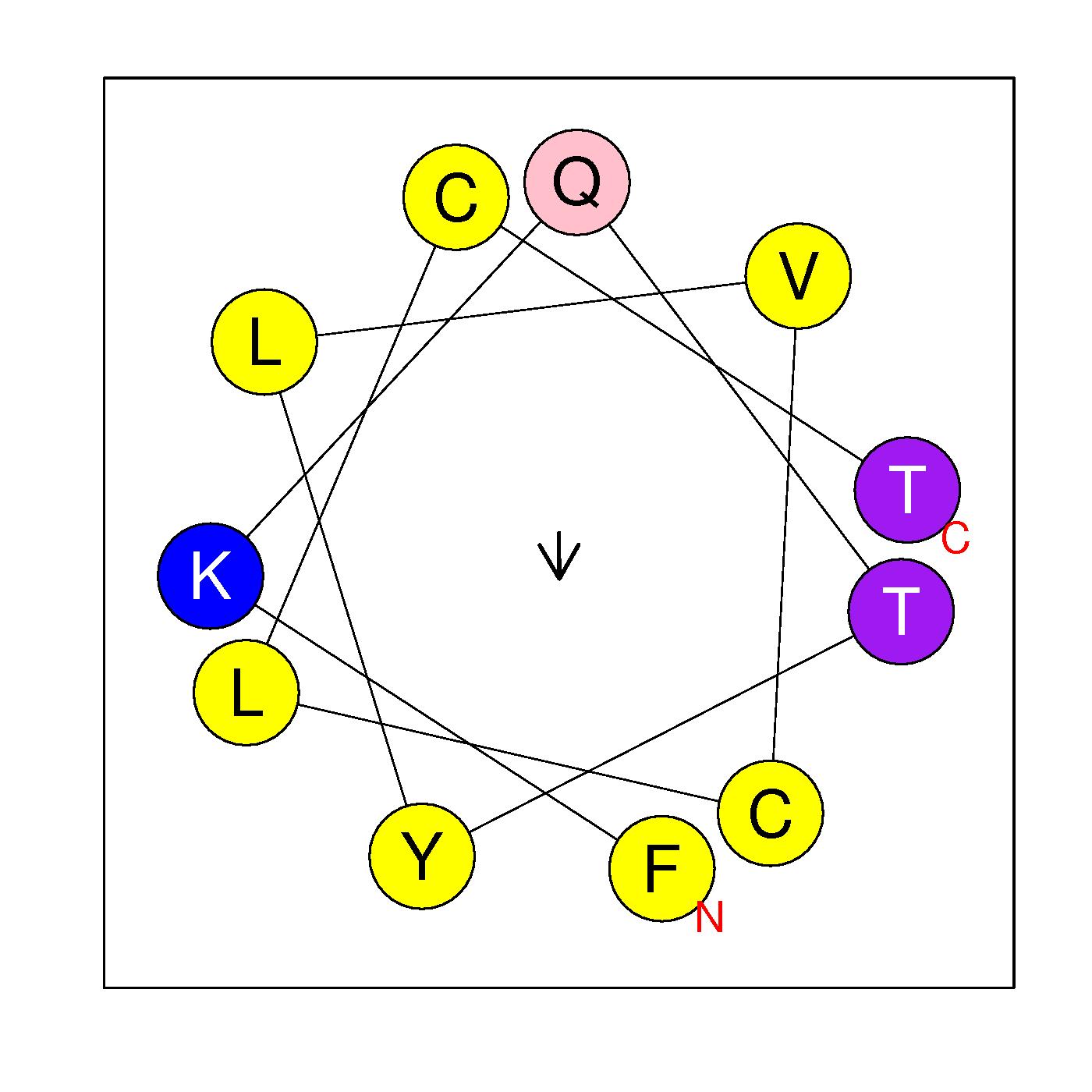
|
|
α12
KLIEEISE
(243-250)
|
H=0.294
Z=-2
|
|
|
α13
CKDALEMICNL
(253-263)
|
H=0.620 μH=0.633
Z=-1
|
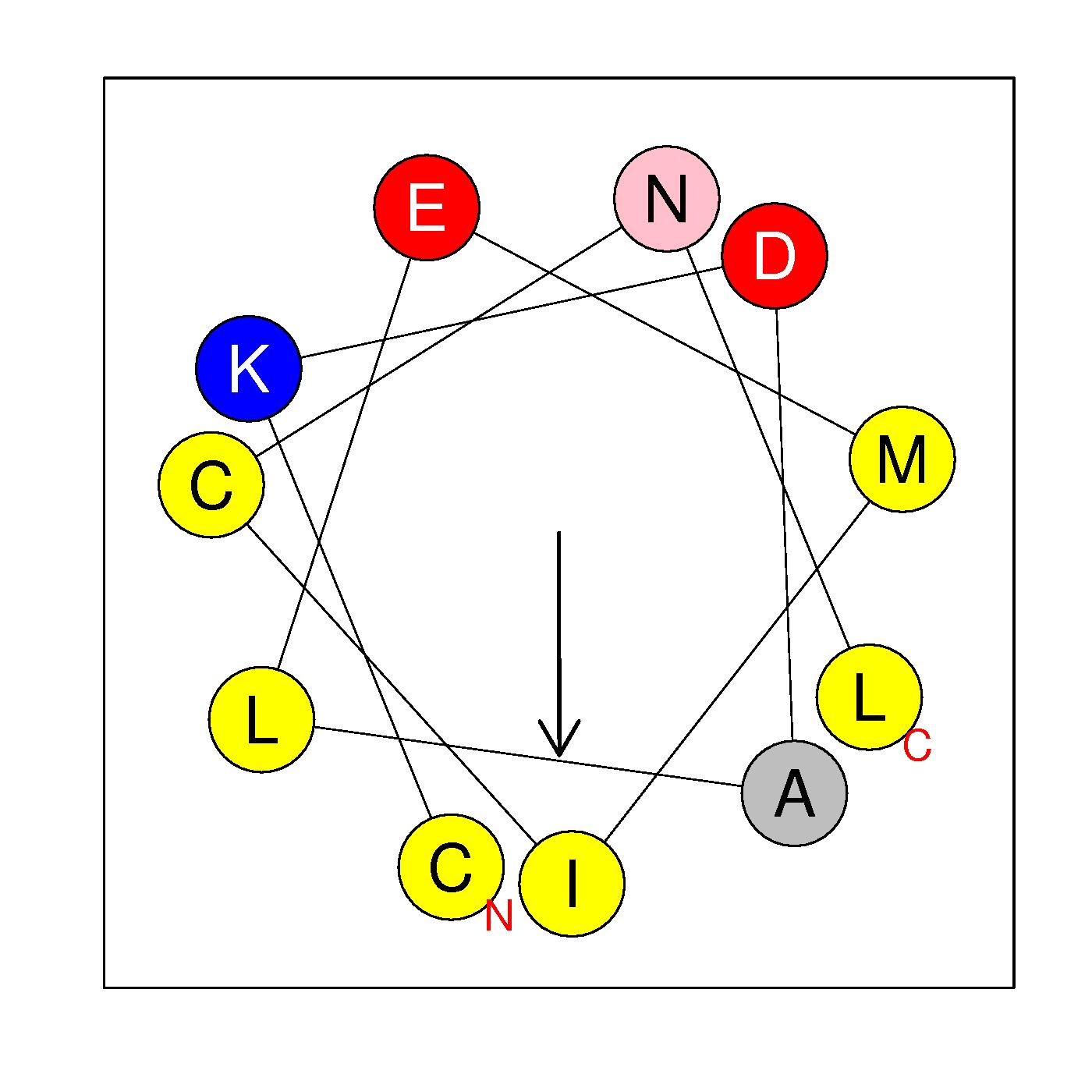
|
|
α14
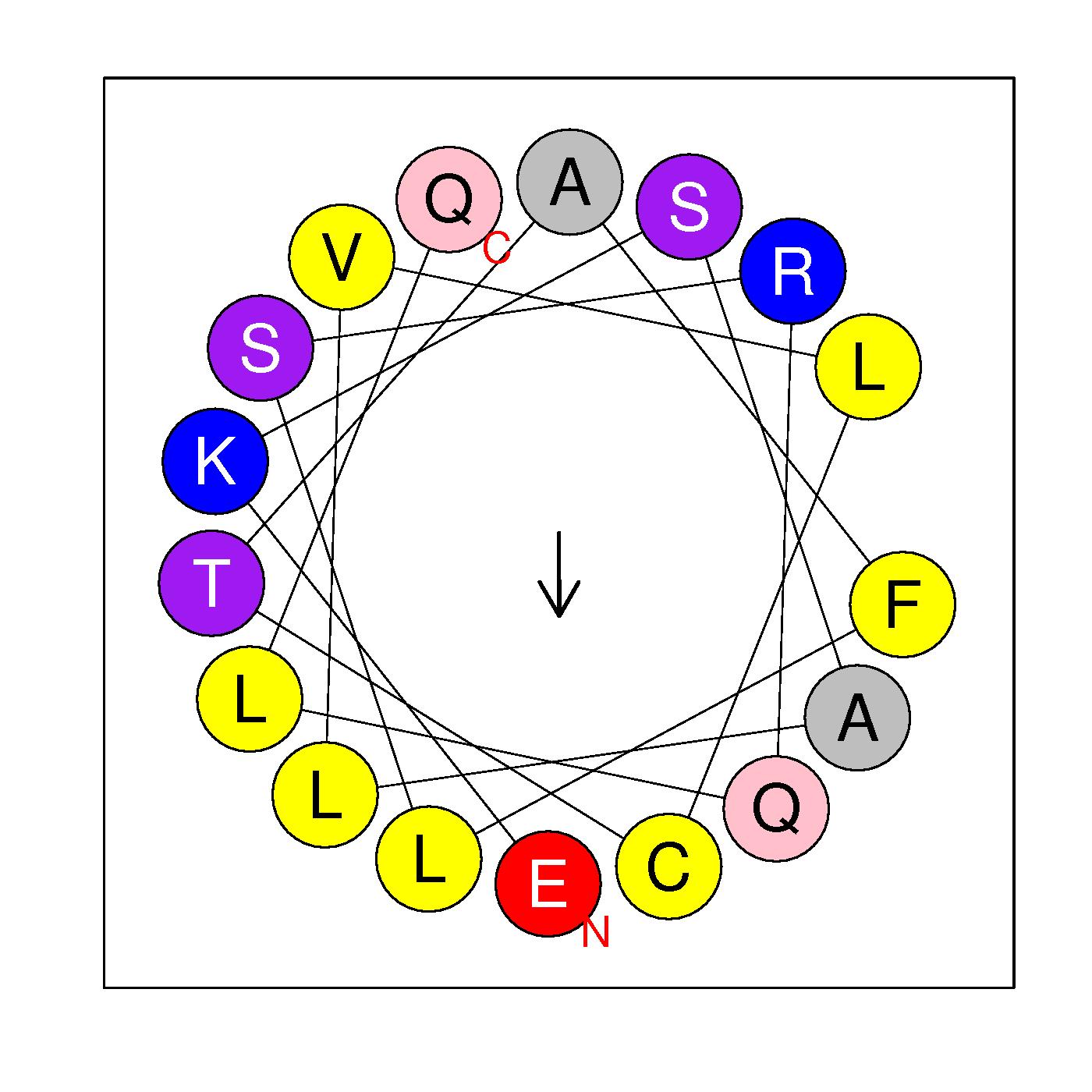
EKSALVLCTAFLSRQLQ
(269-285)
|
H=0.534 μH=0.236
Z=1
|
|
|
α15
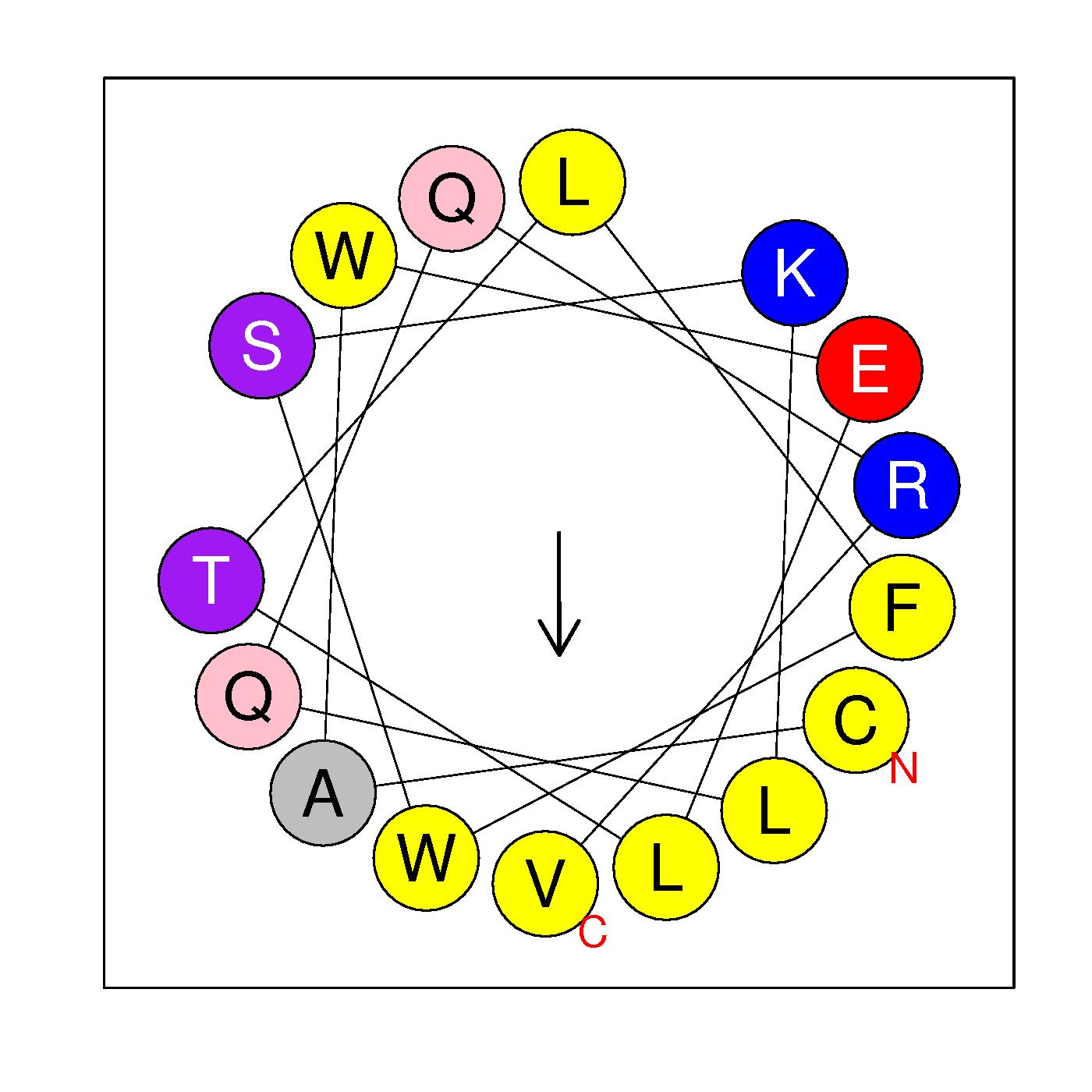
CAWELTLFWSKLQQRV
(291-306)
|
H=0.725 μH=0.347
Z=1
Hydrophobic face L L V W A
|
|
|
α16a
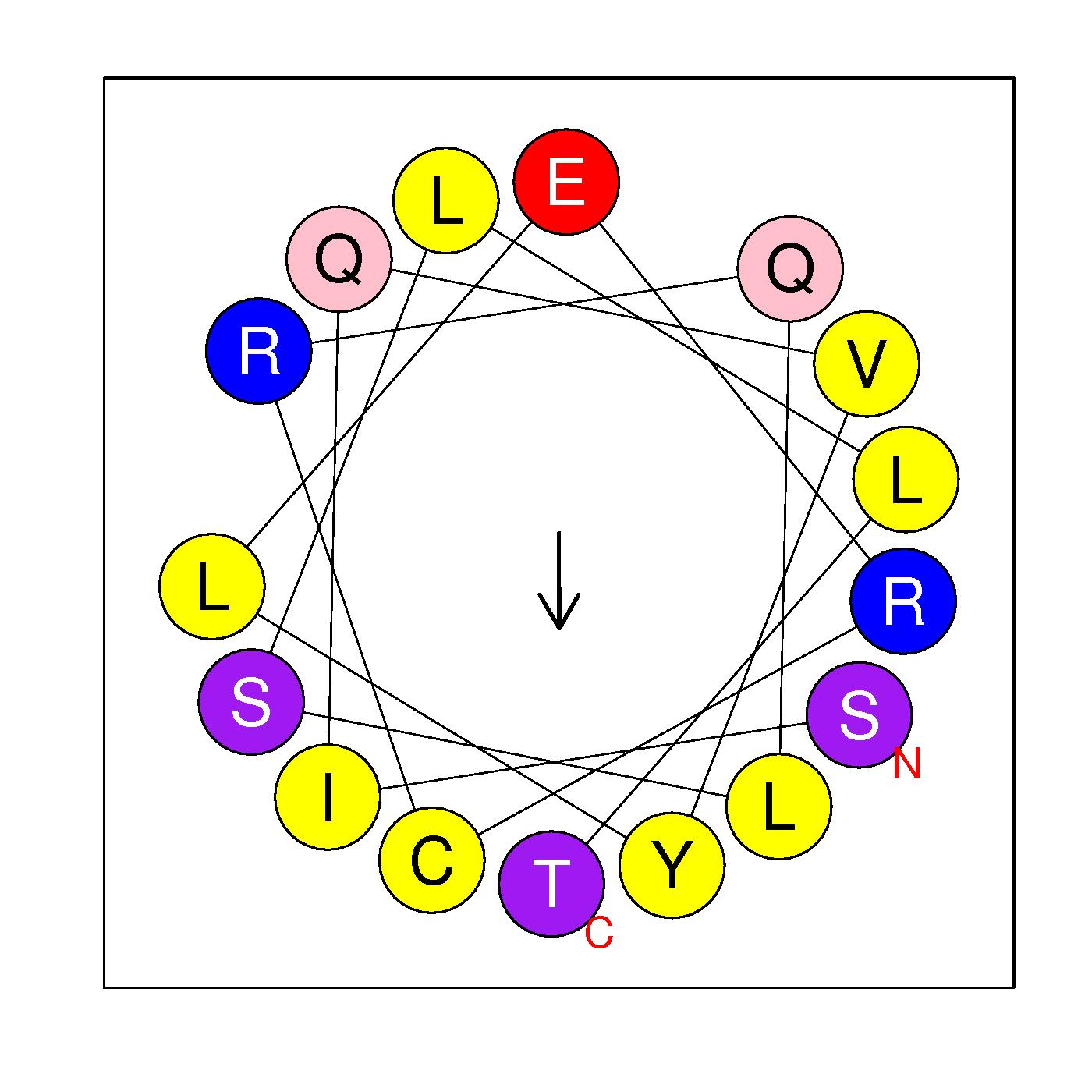
SIQVYLERCRQLSLLT
(309-324)
|
H=0.588 μH=0.272
Z=1
|
|
|
α16b
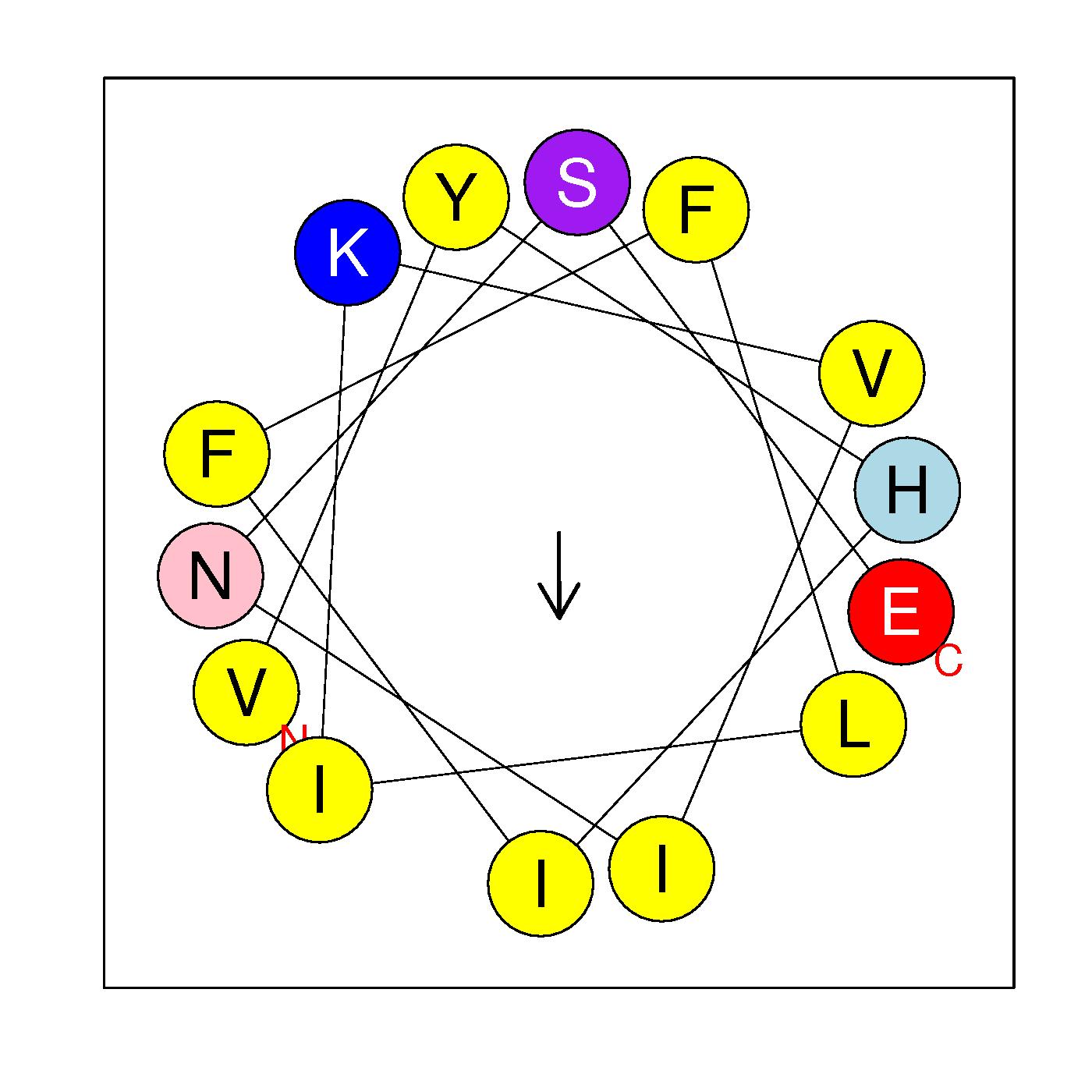
VYHIFFLIKVINSE
(327-340)
|
H=0.853 μH=0.242
Z=0
|
|
|
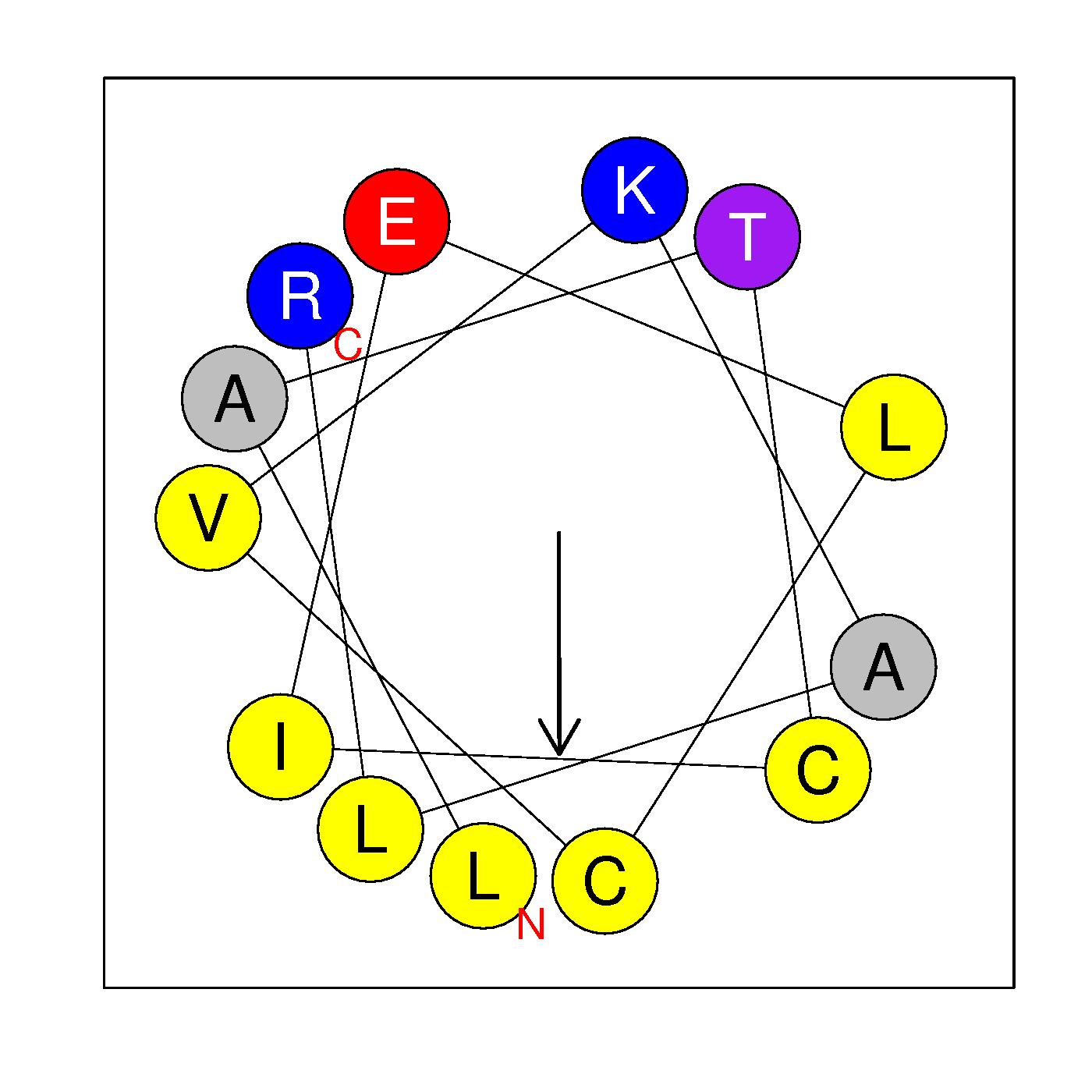
α16c
LATCIELCVKALR
(346-358)
|
H=0.726 μH=0.630
Z=1
|
|
|
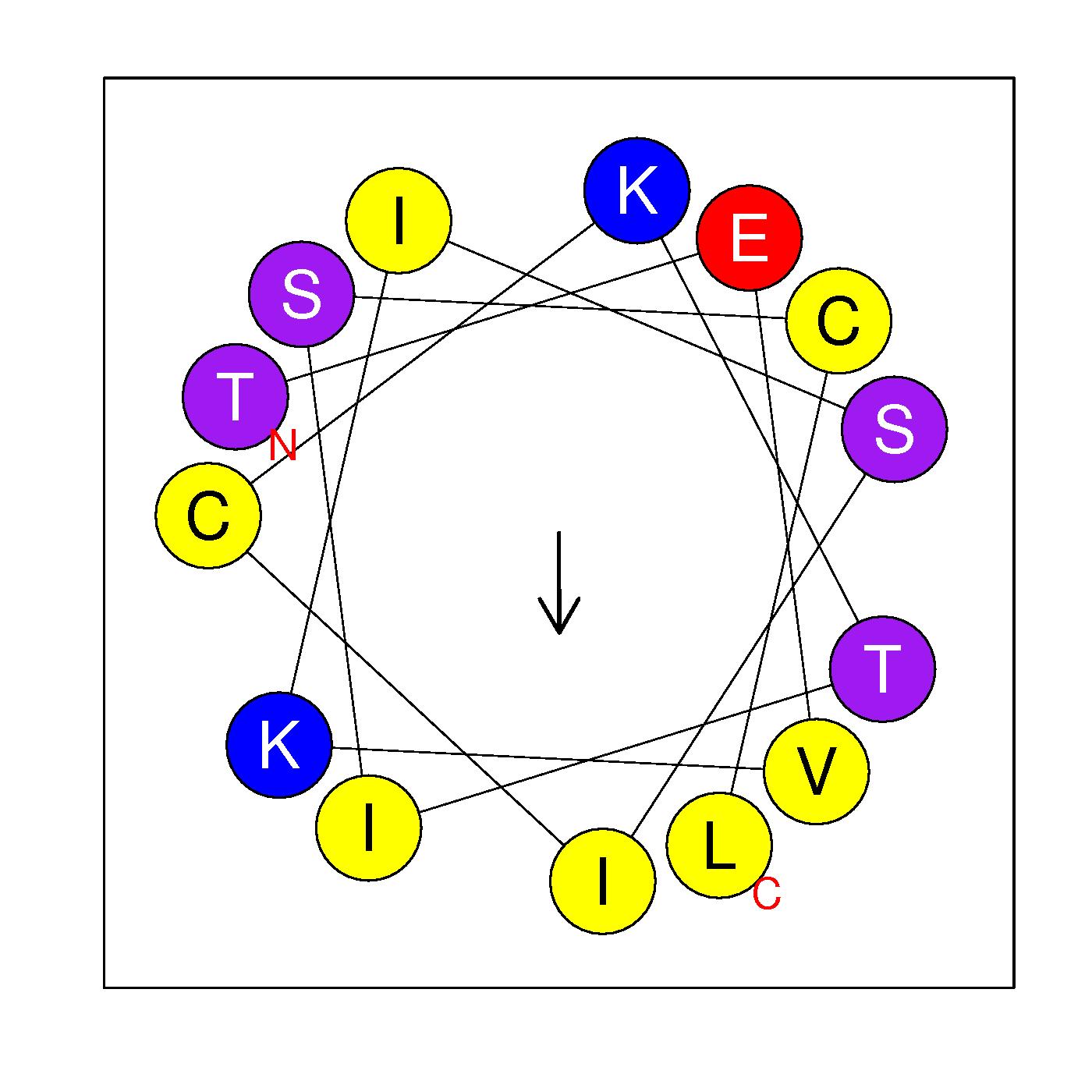
α17
TEVKISICKTISCL
(365-378)
|
H=0.659 μH=0.284
Z=1
|
|
|
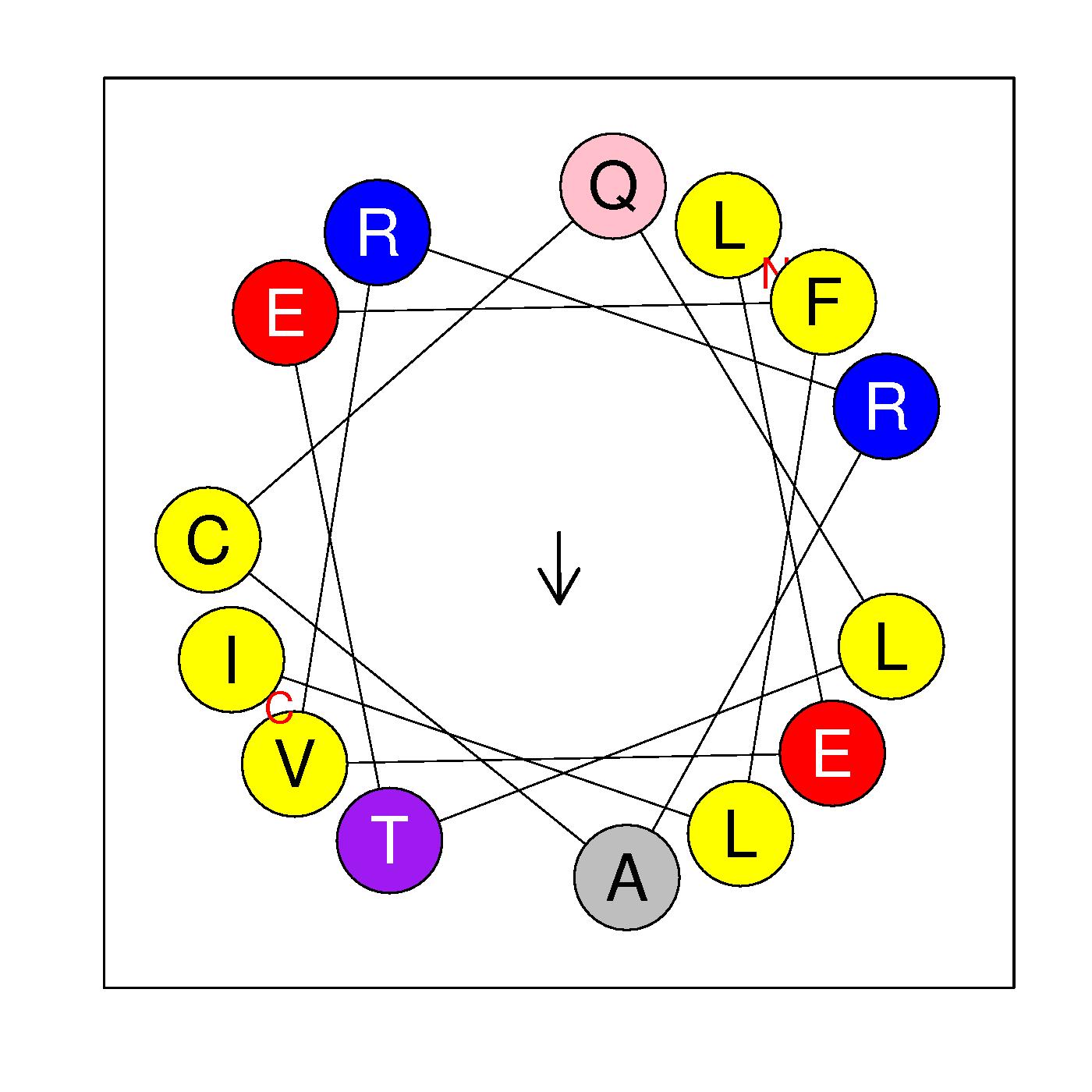
α18
LEVKRACQLSEFLI
(383-396)
|
H=0.587 μH=0.180
Z=0
|
|
|
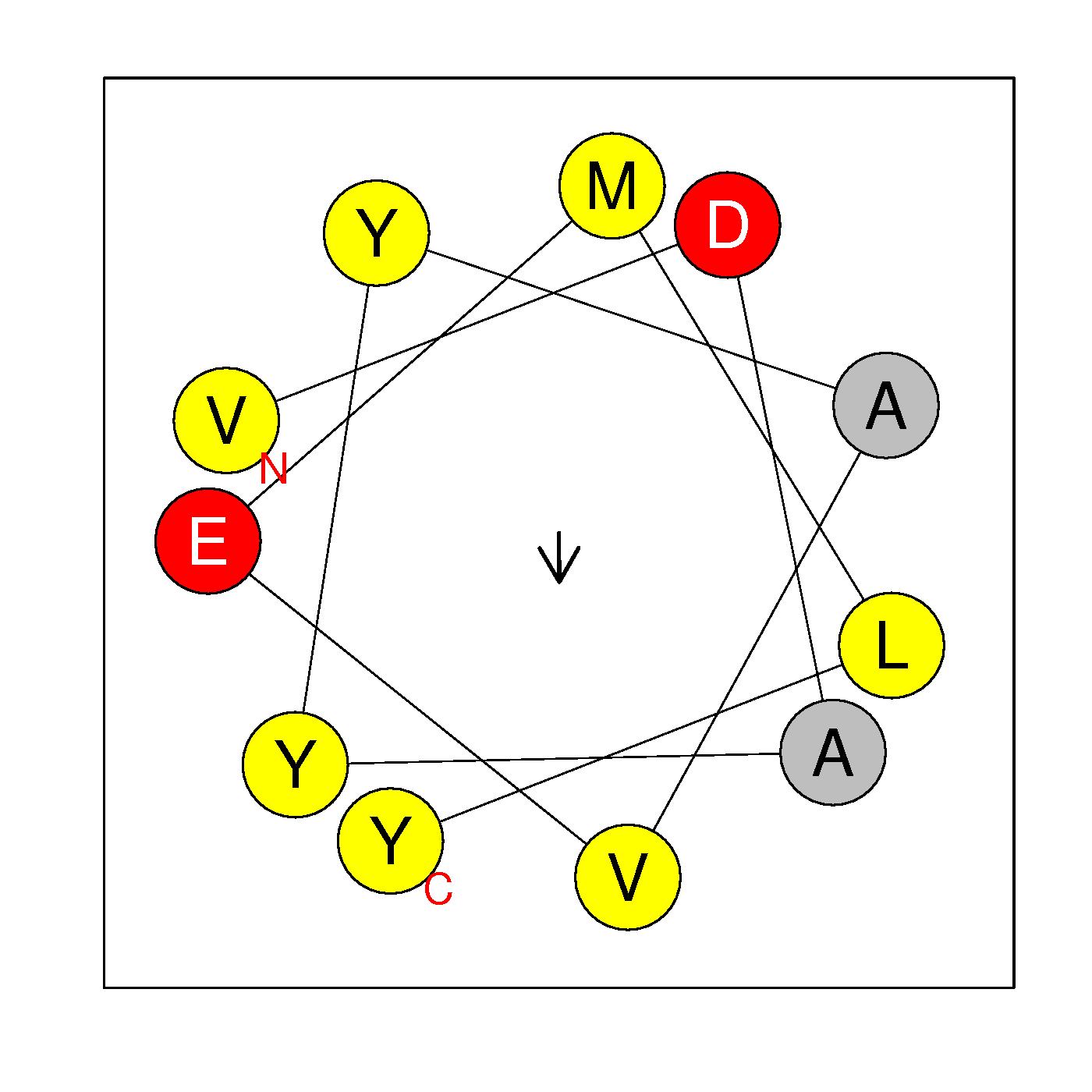
α19
VDAYYAVEMLY
(400-410)
|
H=0.678 μH=0.139
Z=-2
|
|
|
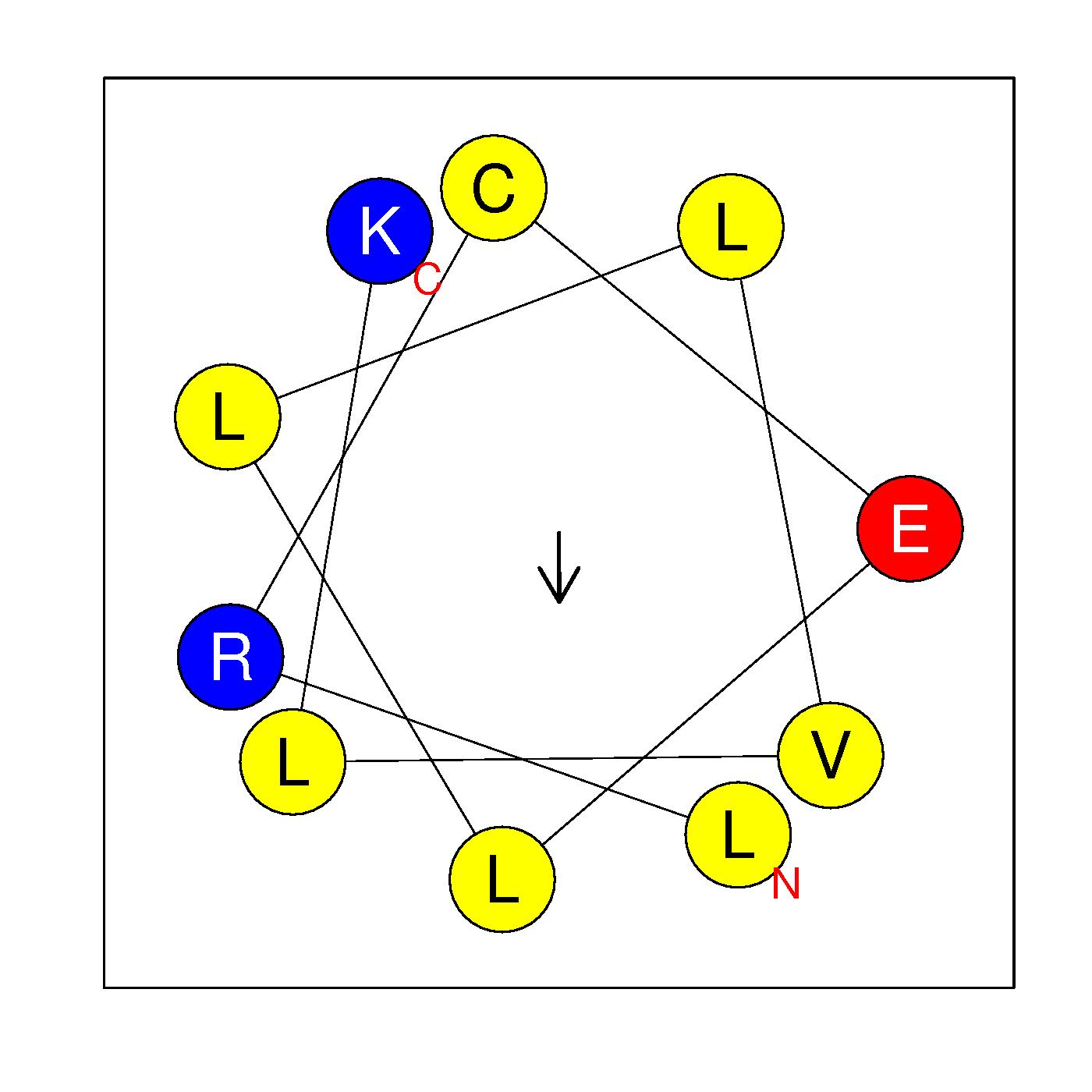
α20
NSLRCELLLVLK
(426:437)
Ambiguous
|
H=0.862 μH=0.197
Z=1
|
|
|
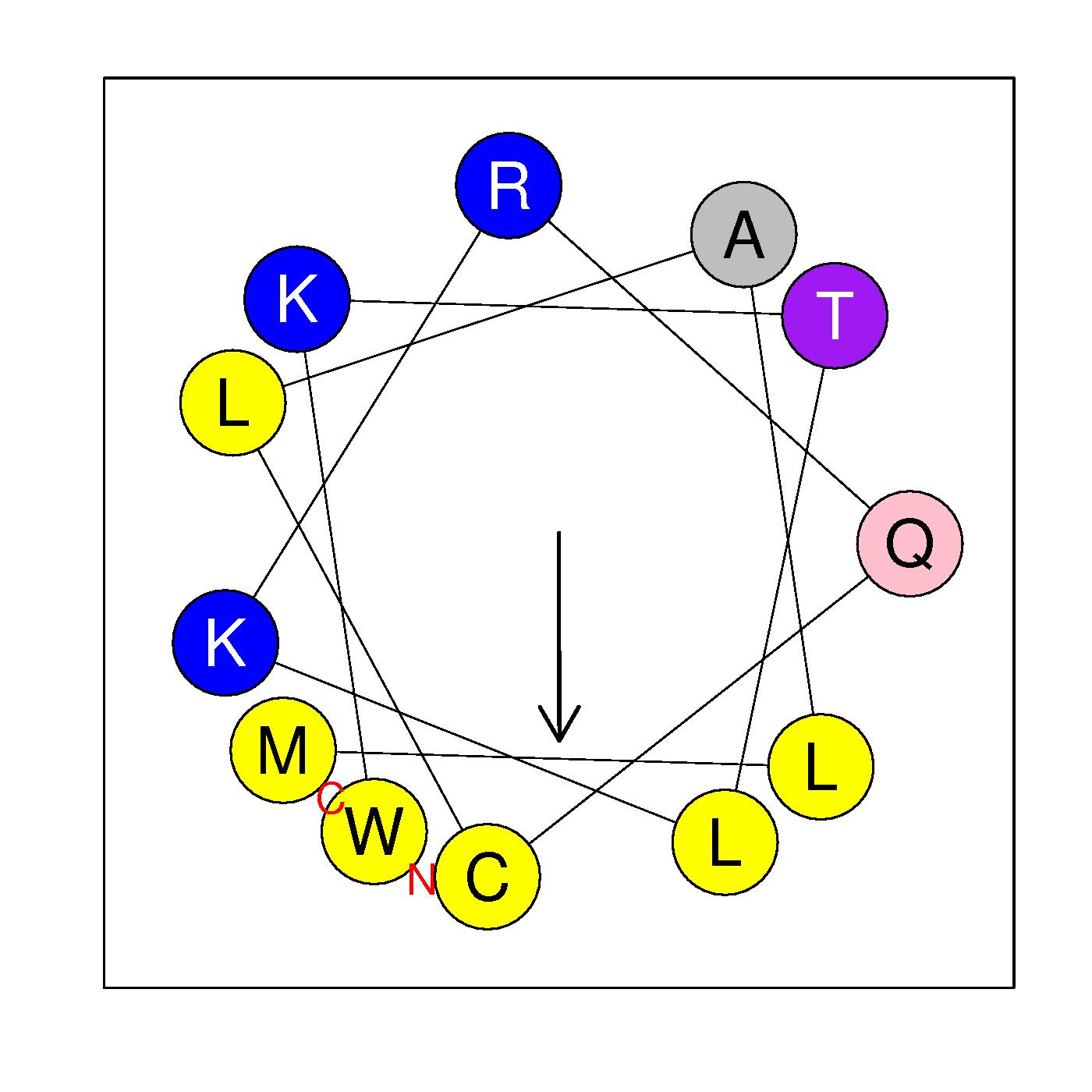
α21a
WKTLKRQCLALM
(449:460)
|
H=0.623 μH=0.592
Z=3
|
|
|
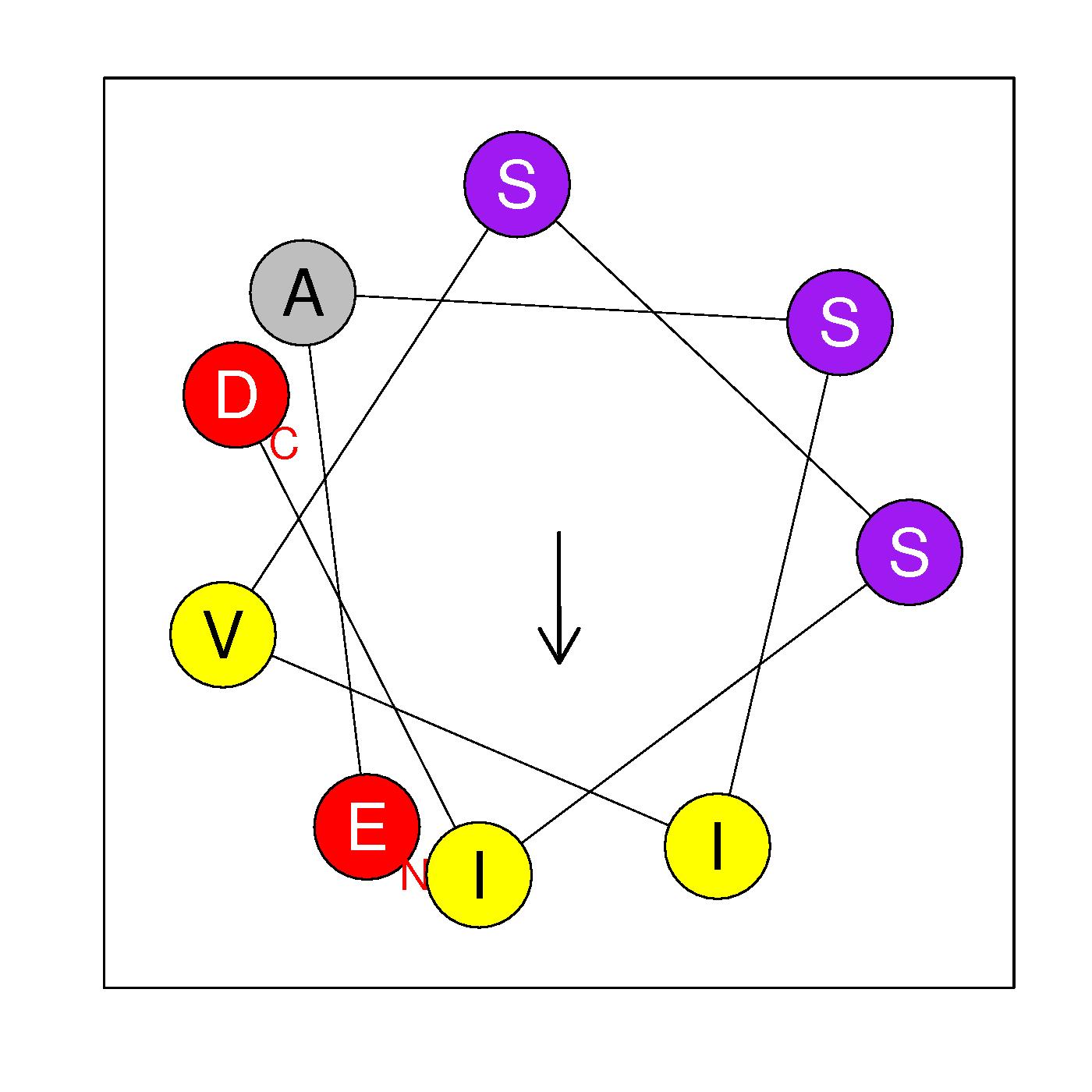
α21b
EASIVSSID
(463:471)
Ambiguous
|
H=0.400 μH=0.370
Z=-2
|
|
|
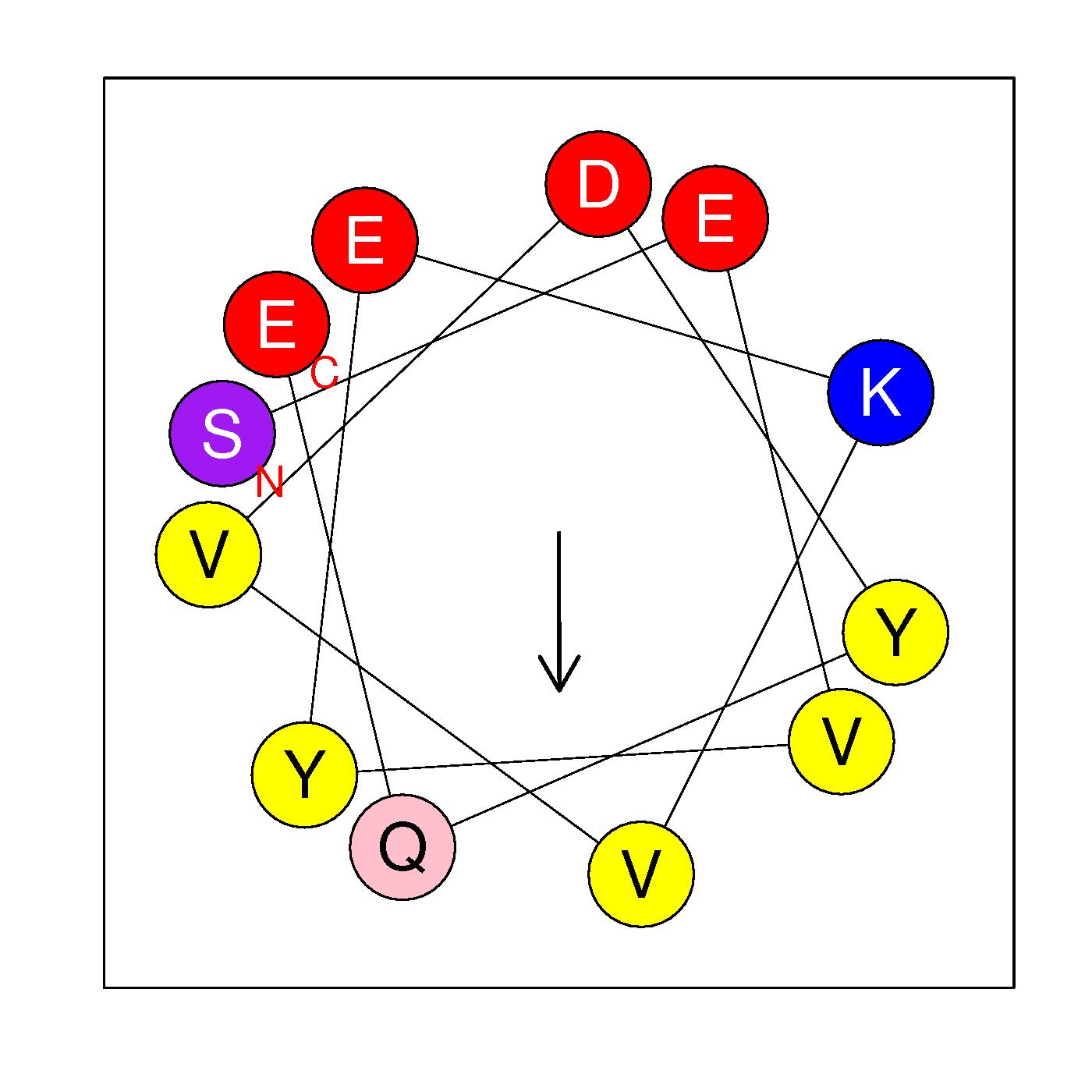
α22
SEVYEKVVDYQE
(476:487)
|
H=0.137 μH=0.448
Z=-3
|
|
|
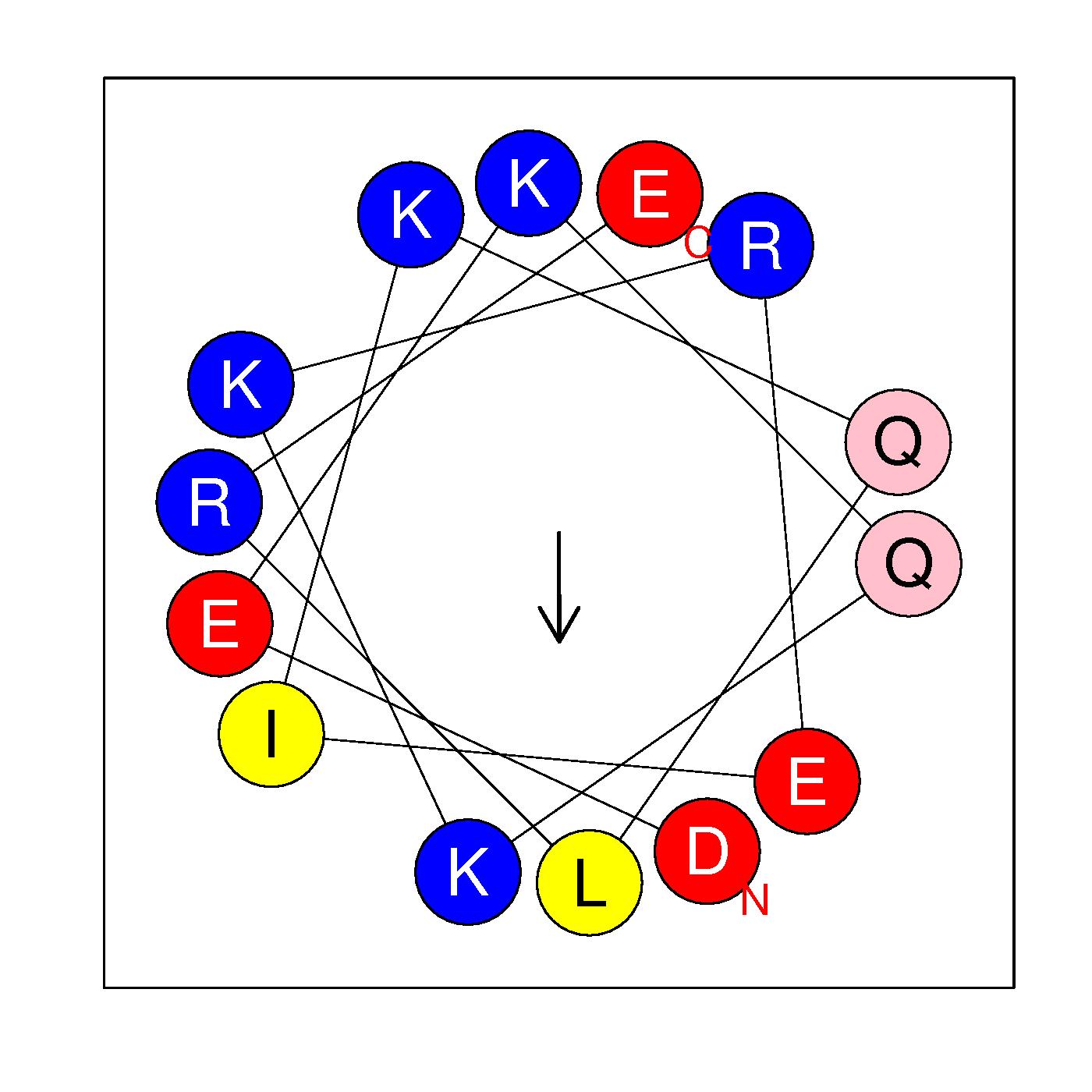
α23
DEKQKKREIKQLRE
(513:526)
|
H=-0.401 μH=0.309
Z=2
|
|
|
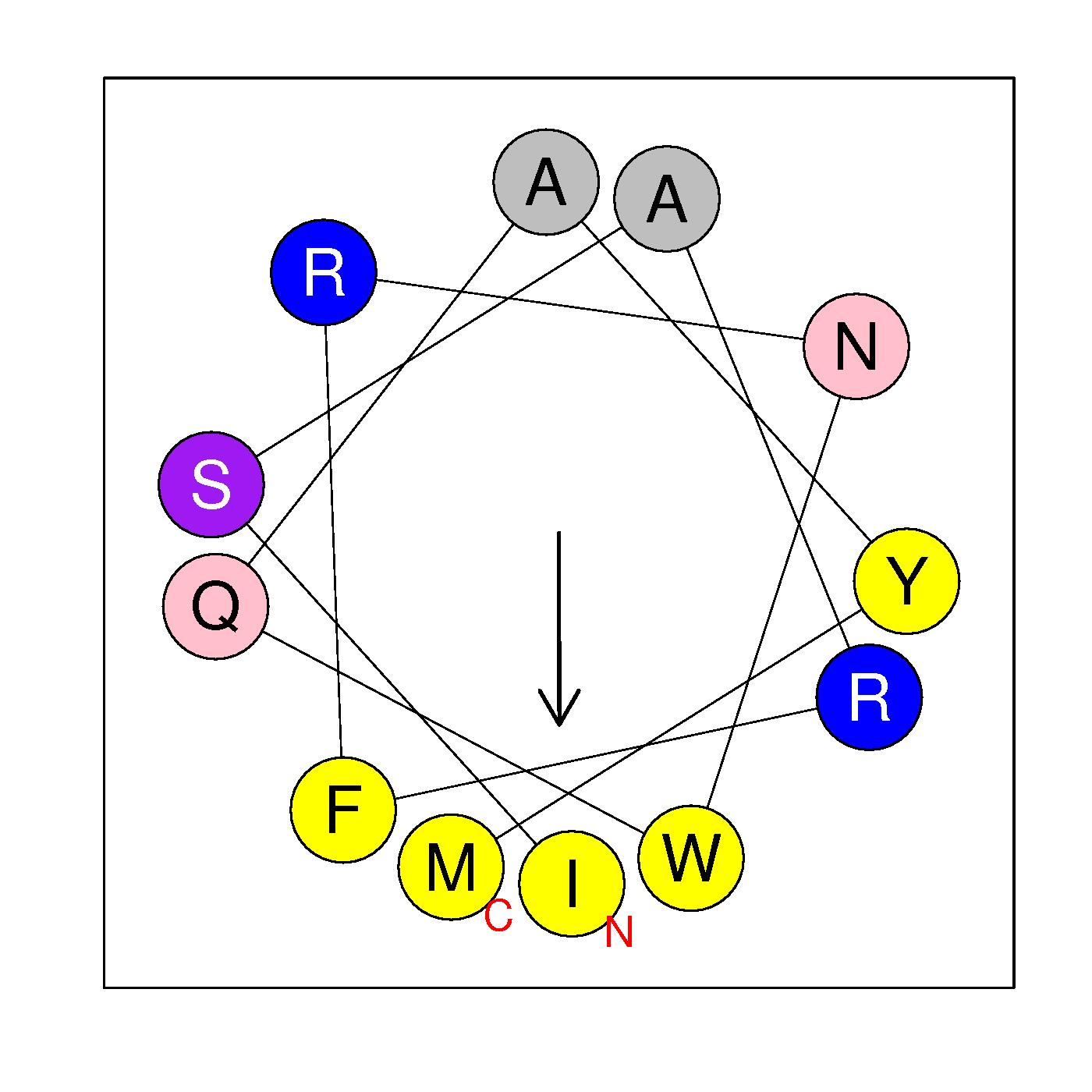
α24
ISARFRNWQAYM
(530:541)
|
H=0.481 μH=0.546
Z=2
|
|
