 |
HOME CONSERVATION C-TERMINAL ANALYSES DNA AND RNA SEQUENCES RESEARCH QUESTIONS RESULTS RESOURCES REFERENCES |
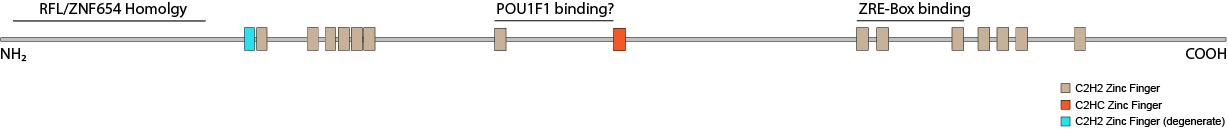
Predicting Zinc Fingers Three pieces of prediction software show 15-16 zinc fingers in ZNF292. The website zf.princeton.edu from the Singh lab predicts 16 with varying scores numbered at 1-16 in the table below. SMART also calls out 16 but lists ZF9 as being degenerate. Uniprot (O60281 · ZN292_HUMAN) does not recognize ZF1. It identifies all others with UniRule PROSITE-ProRule: PRU00042 but also lists ZF9 as degenerate. ZF1 lacks the (F/Y)XC start beginning with QYC instead. ZF9 looks very much like a C2HC zinc finger as is found in FOGs (Friends of GATA). These zinc fingers are thought to bind to a CCCC zinc finger in GATA proteins.
Low Complexity and Disordered Regions
Coiled Coil Predictions Multicoil-2 Prediction Multicoil 2 (http://cb.csail.mit.edu/cb/multicoil2/cgi-bin/multicoil2.cgi) predicts a C-teminal paired coiled coil with a probability of 0.999 stretching from residue 2418 - 2544. SQHRNLLIVFKRCCNSQVKETSEQEGAKNDVKDSDTCVSESNDNSRTTATVSQKEVEKN EKDEMDELTELFITKLINEDSTSETQANTSSNVSNDFQEDNLCQSERQKASNLKRVNKE KNVSQNKKRKVEKA Full results from Multicoil 2 COIL Prediction COIL prediction (https://embnet.vital-it.ch/cgi-bin/COILS_form_parser) from residue 2523-2550 with a probability of 0.841 for 21 aa window or 0.376 for 28 aa window. 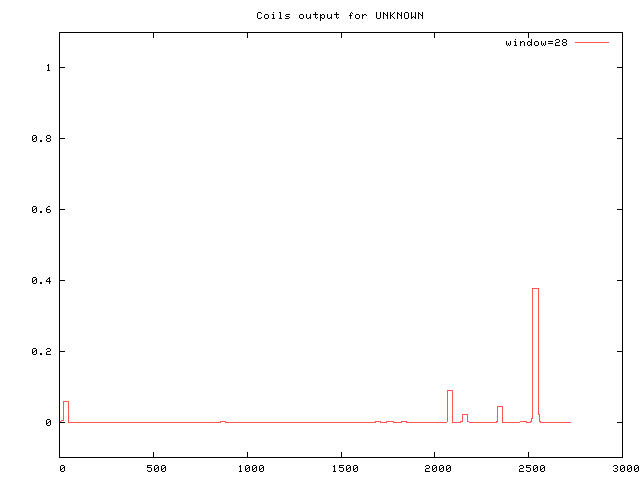
RQKASNLKRVNKEKNVSQNKKRKVEKA Full results from COIL Nuclear Localization Signal
cNLS Mapper Results (6+)
429 RCELLLVLKTQWPFDPEFWDWKTLKRQCL 7.8 bipartite * 2309 EKSKSKHRGTKHSRCGKEGIKMPKTKRKKKN 6.4 bipartite 2316 RGTKHSRCGKEGIKMPKTKRKKKN 10.2 bipartite * 2331 PKTKRKKKN 7 2524 RQKASNLKRVNKEKNVSQNKKRKVE 8.4 bipartite * 2540 SQNKKRKVE 7.5
NucPred Results (Overall score 0.97)
517 KKR 1628 SKRRKKVAP 2090 EEKKRKKPV 2330 MPKTKRKKKNNLENK 2540 NKKRK References |