Playing with Fingers
Canonical forms for DNA binding zinc finger:
Form 1: (F/Y)XCX2-5CX3(F/Y)X5ψX2HX3-5H
Form 2: X2CX2,4CX12H-X3-5H
ZF1 (542-563) SMART E=55.4 zps=2.2 Not in UniProt HMER, non-canonical
QYCX2CX3FX5ψXHX3H
1
■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. QYCVLCDKEFLGHRIVRHAQKH
100% QYClLCDKEFLGHRIVRHAQKH
90% QYCVLCDKEFLGHRIVRHAQKH
80% QYCVLCDKEFLGHRIVRHAQKH
70% QYCVLCDKEFLGHRIVRHAQKH
ph0hh0-+-hh0++hh++hp++
QYCLLCDKEFLGHRIVRHAQKH (Mouse)
***:******************
QYCVLCDKEFLGHRIVRHAQKH (Xenopus Laevis)
**********************
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Asp548Asn 1 1 23 1 5.73 22.4
α=0 β=1
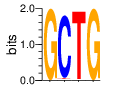
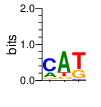
PWM calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.000 0.005 0.005 0.000
c 0.000 0.973 0.005 0.000
g 1.000 0.000 0.000 1.000
t 0.000 0.022 0.989 0.000
Ent= 0.000 0.200 0.098 0.000
|
|
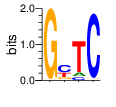
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.000 0.018 0.106 0.005
c 0.000 0.503 0.088 0.980
g 0.999 0.168 0.001 0.014
t 0.001 0.311 0.805 0.001
Ent= 0.014 1.558 0.915 0.161
|
|
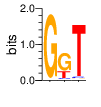
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.009 0.017 0.013
c 0.001 0.052 0.038
g 0.978 0.659 0.018
t 0.012 0.272 0.931
Ent= 0.175 1.230 0.461
|
|
Linker 1-2 (564-568, 5aa)
■■■■■
h.s. YKDGI
100% aKDGl
90% YKDGl
80% YKDGI
70% YKDGI
ZF2 (569-591) SMART E=0.101 zps=18.5 Canonical
YXCX2CX3FX5ψX2HX3H
1 *1
■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. YSCPICAKNFNSKETFVPHVTLH
100% YSCPICAppasSK-sFVPHVThH
90% YSCPICApNFNSKEsFVPHVTLH
80% YSCPICAKNFNSKEoFVPHVTLH
70% YSCPICAKNFNSKETFVPHVTLH
hp00h0h+phpp+-phh0+hph+
YSCPICAKNFNSKDSFVPHVTLH (Mouse)
*************::********
YSCPICAQQYSSKENFVPHVTFH (Xenopus Laevis)
*******:::.***.******:*
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Ile573Val 1 0.999 21 1 5.49 13.47
p.Lys581Arg 22 0.906 26 1 5.49 14.54
p.Glu582Asp 1 0.067 45 1 3 10.53
α=2 β=1
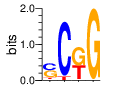
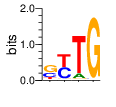
PWM calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.024 0.000 0.009 0.000
c 0.438 0.929 0.001 0.001
g 0.432 0.000 0.568 0.998
t 0.107 0.071 0.422 0.000
Ent= 1.518 0.372 1.062 0.019
|
|
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.018 0.015 0.116 0.009
c 0.308 0.447 0.012 0.017
g 0.500 0.034 0.024 0.968
t 0.175 0.504 0.847 0.006
Ent= 1.567 1.272 0.770 0.252
|
|
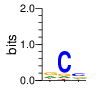
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.293 0.077 0.189
c 0.104 0.756 0.475
g 0.452 0.106 0.217
t 0.151 0.061 0.119
Ent= 1.788 1.181 1.808
|
|
Linker 2-3 (592-671, 80aa)
■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■
h.s. VKQSSKERLAAMKPLRRLGRPPKITTTNENQKTNTVAKQEQRPIKKNSLYSTDFIVFNDNDGSDDENDDKDKSYEPEVIP
100% VKpSsKERLtsMKPLR+lG+PPKhsss..sp+.ss.sppppR.IKKNshY.sDFIVFNDNDtS-D-p.-...........
90% VKpSSKERLAAMKPLRRLGRPPKlssspcNQKssslsKQEQRPIKKNSLYSsDFIVFNDNDGSDDEsDDKDKsY.P-lh.
80% VKQSSKERLAAMKPLRRLGRPPKIosspENQKTNsVsKQEQRPIKKNSLYSTDFIVFNDNDGSDDENDDKDKSYEPEllP
70% VKQSSKERLAAMKPLRRLGRPPKITTTNENQKTNsVuKQEQRPIKKNSLYSTDFIVFNDNDGSDDENDDKDKSYEPEVIP
■■■■■■■■■■■
h.s. VQKPVPVNEFN
100% .........hs
90% lQKPhPVNEFs
80% VQKPlPVNEFN
70% VQKPVPVNEFN
ZF3 (681-705) SMART E=0.352 zps=23.5 Canonical
FXCX4CX3FX5ψX2HX3H
1 1 1 1 2
■■■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. FNCPVTFCKKGFKYFKNLIAHVKGH
100% hsCPVthCKKGFKYFKNLIAHs+GH
90% FsCPVohCKKGFKYFKNLIAHsKGH
80% FNCPVTFCKKGFKYFKNLIAHVKGH
70% FNCPVTFCKKGFKYFKNLIAHVKGH
hp00hph0++0h+hh+phhh+h+0+
FNCPVTFCKKGFKYFKNLIAHVKGH (Mouse)
*************************
FACPVHLCKKGFKYFKNLIAHVRGH (Xenopus Laevis)
* *** :***************:**
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Asn682Ser 1 0.109 46 1 4.86 6.168
p.Phe687Leu 1 0.001 22 1 5.96 2.918
p.Lys689Arg 1 0.33 26 1 5.96 8.586
p.Phe695Ser 1 1 155 1 5.96 18
p.Gly704Ala 2 0.034 60 1 5.07 0.932
α=2 β=3
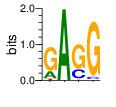
PWM calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.223 1.000 0.000 0.007
c 0.002 0.000 0.266 0.054
g 0.722 0.000 0.705 0.937
t 0.053 0.000 0.029 0.002
Ent= 1.066 0.002 1.013 0.383
|
|
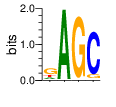
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.184 0.997 0.009 0.012
c 0.056 0.001 0.009 0.880
g 0.527 0.001 0.972 0.096
t 0.233 0.001 0.009 0.011
Ent= 1.658 0.037 0.228 0.639
|
|
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.237 0.733 0.005
c 0.639 0.019 0.054
g 0.084 0.103 0.323
t 0.040 0.145 0.618
Ent= 1.391 1.179 1.219
|
|
Linker 3-4 (706-721)
■■■■■■■■■■■■■■■
h.s. DNEDAKRFLEMQSKK
100% ss--ApRFLEhQSKK
90% DsE-AKRFLEMQSKK
80% DNEDAKRFLEMQSKK
70% DNEDAKRFLEMQSKK
ZF4 (722-744) SMART E=0.153 zps=17.5 Non-canonical (but V is smaller hydrophobic aa)
VXCX2CX3FX5ψX2HX3H
1 1 1 5 1
■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. VICQYCRRHFVSVTHLNDHLQMH
100% VlCQYCRRpFVSlsHLNDHLQMH
90% VICQYCRRHFVSVTHLNDHLQMH
80% VICQYCRRHFVSVTHLNDHLQMH
70% VICQYCRRHFVSVTHLNDHLQMH
hh0ph0+++hhphp+hp-+hph+
VICQYCRRHFVSVTHLNDHLQMH (Mouse)
***********************
VVCQYCRRQFVSLAHLNDHLQMH (Xenopus Laevis)
*:******:***::*********
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Gln725Arg 1 0.996 43 1 5.97 18.21
p.Arg729Gln 1 1 43 0.99 5.97 22.9
p.Val734Ile 1 0.959 21 1 5.97 17.7
p.Asn738Ser 5 1 46 0.999 4.82 15.7
p.Gln742Arg 1 0.852 43 1 5.97 15.02
α=3 β=2
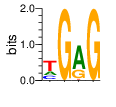
PWM calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.215 0.000 0.701 0.000
c 0.214 0.000 0.000 0.000
g 0.006 1.000 0.267 1.000
t 0.565 0.000 0.032 0.000
Ent= 1.464 0.002 1.031 0.001
|
|
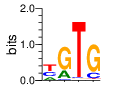
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.217 0.244 0.008 0.018
c 0.316 0.047 0.029 0.168
g 0.008 0.707 0.015 0.777
t 0.460 0.002 0.947 0.037
Ent= 1.573 1.074 0.373 0.995
|
|
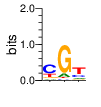
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.078 0.116 0.129
c 0.586 0.039 0.224
g 0.094 0.798 0.091
t 0.242 0.047 0.556
Ent= 1.555 1.011 1.649
|
|
Linker 4-5 (745-749)
■■■■■
h.s. CGSKP
100% CGspP
90% CGSKP
80% CGSKP
70% CGSKP
ZF5 (750-774) SMART E=1.23 zps=8.9 Canonical
YXCX4CX3FX5ψX2HX3H
1 1 1
■■■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. YICIQMKCKAGFNSYAELLTHRKEH
100% YICIQhKCKAuFpoYA-LLsHR+EH
90% YICIQMKCKAGFNSYAELLsHRKEH
80% YICIQMKCKAGFNSYAELLTHRKEH
70% YICIQMKCKAGFNSYAELLTHRKEH
hh0hph+0+h0hpphh-hhp+++-+
YICIQMKCKAGFNSYAELLAHRKEH (Mouse)
*******************:*****
YICIQMKCKASFETYADLLSHRKEH (Xenopus Laevis)
**********.*::**:**:*****
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Ile753Thr 1 1 89 1 5.76 16.77
p.Lys758Gln 1 1 53 1 5.76 17.19
p.Thr769Ser 1 0.007 58 1 5.76 4.553
α=1 β=2
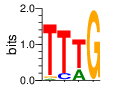
PWM calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.034 0.001 0.278 0.000
c 0.000 0.136 0.001 0.000
g 0.043 0.003 0.001 0.993
t 0.924 0.860 0.719 0.007
Ent= 0.466 0.608 0.879 0.059
|
|
PWM calculated using zf.princeton.edu with Polynomial SVM
|
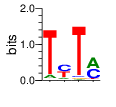
base 1 2 3 4
a 0.079 0.124 0.021 0.484
c 0.014 0.455 0.031 0.435
g 0.017 0.026 0.028 0.011
t 0.891 0.394 0.920 0.070
Ent= 0.621 1.559 0.529 1.367
|
|
PWM calculated using zf.princeton.edu with B1H RFR
|
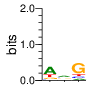
base 1 2 3
a 0.564 0.440 0.111
c 0.071 0.206 0.119
g 0.138 0.197 0.636
t 0.227 0.157 0.134
Ent= 1.615 1.872 1.521
|
|
Linker 5-6 (775-778)
■■■■
h.s. QVFR
100% .VFR
90% QVFR
80% QVFR
70% QVFR
ZF6 (779-803) SMART E=10.8 zps=15.9 Non-canonical (but A is smaller hydrophobic aa)
AXCX4CX3FX5ψX2HX3H
1 1
■■■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. AKCMFPKCGRIFSEAYLLYDHEAQH
100% A+ChFPKCGRlFStAYhLaDHEAQH
90% AKChFPKCGRIFSpAYLLYDHEAQH
80% AKCMFPKCGRIFSEAYLLYDHEAQH
70% AKCMFPKCGRIFSEAYLLYDHEAQH
h+0hh0+00+hhp-hhhhh-+-hp+
AKCLFPKCGRIFSQAYLLYDHEAQH (Mouse)
***:*********:***********
ARCMFPKCGRIFSAAYMLFDHEAQH (Xenopus Laevis)
*:*********** **:*:******
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Ala801Val 1 1 64 1 5.76 17.96
p.His803Arg 1 1 29 1 5.76 16.51
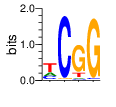
α=2 β=0
PWM calculated using zf.princeton.edu with Expanded Linear SVM
|
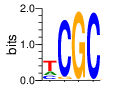
base 1 2 3 4
a 0.215 0.001 0.003 0.000
c 0.214 0.947 0.005 0.985
g 0.006 0.051 0.991 0.015
t 0.565 0.001 0.002 0.000
Ent= 1.464 0.316 0.090 0.110
|
|
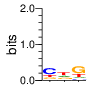
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.230 0.001 0.045 0.005
c 0.230 0.971 0.031 0.027
g 0.008 0.024 0.785 0.963
t 0.532 0.004 0.138 0.005
Ent= 1.517 0.213 1.026 0.273
|
|
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.061 0.234 0.096
c 0.541 0.078 0.088
g 0.187 0.231 0.552
t 0.211 0.457 0.264
Ent= 1.652 1.781 1.614
|
|
Linker 6-7 (804-806)
■■■
h.s. YNT
100% YNT
90% YNT
80% YNT
70% YNT
ZF7 (807-831) SMART E=0.000369 zps=29.8 HMMER=3.8e-07 Canonical
YXCX4CX3YX5ψX2HX3H
2 *1 1 3
■■■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. YTCKFTGCGKVYRSQGELEKHLDDH
100% aTC+hssCGKlY+SQ.ph-KH.ptH
90% YTCKFTGCGKVYRSQsEhEKHl--H
80% YTCKFTGCGKVYRSQuELEKHL-DH
70% YTCKFTGCGKVYRSQuELEKHLDDH
hp0+hp000+hh+pp0-h-++h--+
YTCKFTGCGKVYRSQSEMEKHQDGH (Mouse)
***************.*:*** *.*
FTCKYVGCGKIYHSQLQLEKHLSEH (Xenopus Laevis)
:***:.****:*:** :*****.:*
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Gly813Asp 2 0.001 94 1 3.99 5.806
p.Arg819Cys 4 1 180 1 5.76 15.96
p.Arg819His 6 0.477 29 1 5.76 10.78
p.Ser820Pro 1 1 74 1 5.76 17.67
p.Glu823Gly 1 0.999 98 1 5.76 16
p.Asp829His 1 0.808 88 1 5.67 14.17
p.Asp829Asn 2 0.196 23 1 5.62 13.09
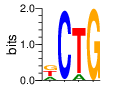
α=4 β=1
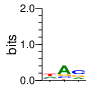
PWMs calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.250 0.002 0.122 0.000
c 0.002 0.993 0.000 0.000
g 0.374 0.002 0.002 1.000
t 0.374 0.002 0.876 0.000
Ent= 1.581 0.072 0.558 0.000
|
|
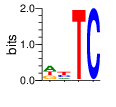
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.413 0.193 0.002 0.000
c 0.004 0.324 0.002 0.999
g 0.289 0.060 0.001 0.001
t 0.293 0.423 0.995 0.000
Ent= 1.599 1.753 0.057 0.009
|
|
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.296 0.596 0.229
c 0.182 0.142 0.454
g 0.094 0.186 0.269
t 0.428 0.075 0.049
Ent= 1.812 1.578 1.726
|
|
ZF8 (1098-1123) SMART E=0.0767 zps=21 Canonical
FXCX4CX3YX5ψX2HX4H
1 14
■■■■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. FSCQVEGCTRTYNSSQSIGKHMKTAH
100% FpCpVEGCTR.YNSsQSIGKHMKsAH
90% FsCQVEGCTRoYNSSQSIGKHMKTAH
80% FSCQVEGCTRTYNSSQSIGKHMKTAH
70% FSCQVEGCTRTYNSSQSIGKHMKTAH
hp0ph-00p+phppppph0++h+ph+
FSCQVEGCTRTYNSSQSIGKHMKTAH (Mouse)
**************************
FKCSVEGCTRIYNSVQSIGKHMKTAH (Xenopus Laevis)
*.*.****** *** ***********
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Gly1104Glu 1 1 98 1 5.55 18.29
p.Ser1114Gly 1 0.997 56 1 5.55 22.2
p.Ile1115Ser 4 0.999 142 1 5.55 21
α=2 β=1
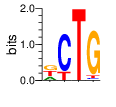
PWM calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.250 0.000 0.001 0.007
c 0.002 0.811 0.000 0.036
g 0.374 0.000 0.000 0.893
t 0.374 0.188 0.998 0.064
Ent= 1.581 0.707 0.018 0.622
|
|
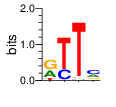
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.352 0.001 0.019 0.297
c 0.002 0.248 0.018 0.479
g 0.509 0.000 0.021 0.144
t 0.137 0.751 0.942 0.080
Ent= 1.435 0.824 0.412 1.722
|
|
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.207 0.406 0.184
c 0.336 0.352 0.448
g 0.103 0.198 0.208
t 0.354 0.045 0.160
Ent= 1.868 1.722 1.862
|

|
ZF9 (1375-1397) SMART E=193 zps=11.7 (degenerate in UniProt) Non-canonical (longer α-helix with terminal C)
Fits Interpro ZF_C2HC_RNF
FXCX3CX3FX5ψX2HX6C
2
■■■■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. FICSRCYRAFTNPRSLGGHLSKRSYC
100% FICSRCaRsFoNPRSLGGHLSKRuhC
90% FICSRCaRsFTNPRSLGGHLSKRSaC
80% FICSRCYRAFTNPRSLGGHLSKRSYC
70% FICSRCYRAFTNPRSLGGHLSKRSYC
hh0p+0h+hhpp0+ph00+hp++ph0
FICSRCYRAFTNPRSLGGHLSKRSYC (Mouse)
**************************
FICSRCFRAFTNPRSLGGHLSKRAVC
******:****************: *
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Pro1387His 2 1 77 1 5.32 16.89
α=1 β=0
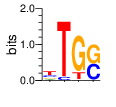
PWM calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.124 0.000 0.013 0.000
c 0.255 0.069 0.001 0.453
g 0.124 0.000 0.815 0.547
t 0.497 0.931 0.171 0.000
Ent= 1.750 0.363 0.765 0.994
|
|
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.206 0.001 0.308 0.002
c 0.216 0.078 0.127 0.801
g 0.206 0.000 0.269 0.193
t 0.373 0.921 0.296 0.005
Ent= 1.947 0.409 1.930 0.765
|
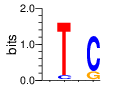
|
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.131 0.150 0.081
c 0.194 0.075 0.846
g 0.027 0.577 0.038
t 0.649 0.198 0.035
Ent= 1.387 1.612 0.846
|
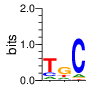
|
Linker 9-10
coiled-coil (1830-1850)
■■■■■■■■■■■■■■■■■■■■■
h.s. VSHKEDQIQEILEGLQKLKLE
100% ...c..ph.-Ih-hlppLpL.
90% h..K-sQl.EILEGLppLKLE
80% h.pK-DQlQEILEGLQKLKLE
70% l.HKEDQIQEILEGLQKLKLE
Suspected Z-Box binding (10-12)
ZF10 (1904-1927) SMART E=0.0767 zps=10.9 Non-canonical
FXCX4CX1YX7ψX2HX4H
1
■■■■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. FVCQNQGCNYSAMTKDALFKHYGKIH
100% FhCQt.sCsYsAMTKDALFKHYuKlH
90% FVCQspGCsYSAMTKDALFKHYuKIH
80% FVCQNQGCNYSAMTKDALFKHYGKIH
70% FVCQNQGCNYSAMTKDALFKHYGKIH
hh0ppp00phphhp+-hhh++h0+h+
FVCQNQGCNYSAMTKDALFKHYGKIH (Mouse)
**************************
FICQETDCTYCAMTKDALFKHYAKVH (Xenopus Laevis)
*:**: .*.*.***********.*:*
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Phe1920Ile 1 0.976 21 0.933 6.17 16.61
α=1 β=0
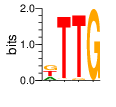
PWM calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.250 0.011 0.000 0.000
c 0.002 0.002 0.000 0.000
g 0.374 0.017 0.035 1.000
t 0.374 0.969 0.965 0.000
Ent= 1.581 0.236 0.218 0.003
|
|
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.263 0.017 0.003 0.000
c 0.001 0.007 0.000 1.000
g 0.466 0.004 0.003 0.000
t 0.270 0.973 0.994 0.000
Ent= 1.540 0.217 0.063 0.004
|
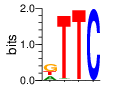
|
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.055 0.262 0.159
c 0.064 0.235 0.289
g 0.393 0.309 0.248
t 0.488 0.195 0.305
Ent= 1.519 1.980 1.960
|
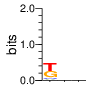
|
Linker 10-11 (1928-1946, 19aa)
■■■■■■■■■■■■■■■■■■■
h.s. QYTPEMILEIKKNQLKFAP
100% .YT.EhIh-IKKpQLKaAP
90% QYTsEMILEIKKpQLKaAP
80% QYTPEMILEIKKNQLKFAP
70% QYTPEMILEIKKNQLKFAP
ZF11 (1947-1972) SMART E=193 zps=27.9 HMMER=2.1e-06 Canonical
FXCX4CX3FX5ψX2HX4H
1 1 1
■■■■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. FKCVVPTCTKTFTRNSNLRAHCQLVH
100% FKCVVsoCsKTFTRNSNLRAHCQ.hH
90% FKCVVsoCsKTFTRNSNLRAHCQLVH
80% FKCVVPTCTKTFTRNSNLRAHCQLVH
70% FKCVVPTCTKTFTRNSNLRAHCQLVH
h+0hh0p0p+php+ppph+h+0phh+
FKCVVPSCTKTFTRNSNLRAHCQLVH (Mouse)
******:*******************
FKCVVPSCTKTFTRNSNLRAHCQSMH (Xenopus Laevis)
******:**************** :*
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Val1951Ile 1 0.997 29 1 6.11 13.62
p.Arg1965Gly 1 1 125 0.998 6.11 15.18
p.Leu1970Phe 1 0.267 22 1 4.35 11.09
α=2 β=1
PWMs calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.223 1.000 0.000 0.000
c 0.002 0.000 0.000 0.168
g 0.722 0.000 0.999 0.813
t 0.053 0.000 0.001 0.018
Ent= 1.066 0.002 0.010 0.782
|
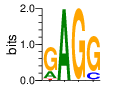 |
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.137 1.000 0.000 0.152
c 0.040 0.000 0.000 0.846
g 0.517 0.000 1.000 0.001
t 0.305 0.000 0.000 0.000
Ent= 1.593 0.004 0.005 0.632
|
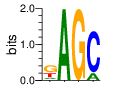
|
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.288 0.884 0.003
c 0.519 0.008 0.026
g 0.023 0.043 0.856
t 0.170 0.065 0.115
Ent= 1.567 0.665 0.711
|
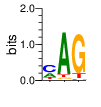
|
Linker 11-12 (1973-2113, 141 aa)
■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■
h.s. HFTTEEMVKLKIKRPYGRKSQSENVPASRSTQVKKQLAMTEENKKESQPALELRAETQNTHSNVAVIPEKQLVEKKSPDK
100% pFT.EpMlKLKlKRsYG++sp.p........p...........p..................p...h.tp...E....p.
90% HFTsEEMVKLKIKRPYGR+oQsEs.sssp..plpp...hh.-scpp...shph....p.sh.p.shl.-p.h.EppsP-p
80% HFTTEEMVKLKIKRPYGRKSQsEs.sssp.sQVK+Q.shsEEsKpE.psshclts.ppsshsslullPEKQlhEKKSP-+
70% HFTTEEMVKLKIKRPYGRKSQsENlousp.sQVK+Q.shTEENK+E.QPslELts.pcsshsNlAVIPEKQLhEKKSP-K
■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■
h.s. TESSLQVITVTSEQCNTNALTNTQTKGRKIRRHKKEKEEKKRKKPVSQSLEFPTRYSPYRP
100% .t.s................tp...KhpK.++.pKEhp-h.thp..pp....stpassY+P
90% .Eps.p.hshs.-p.s.ts.sphp.KsRK.+R++KEKEE++c+pPsspu.EhPs+YSsYRP
80% hEsS.QslslssEQhs.sshoshpsKGRKhRRH+KEKEE++cKKPVopShEhPTRYSPYRP
70% sESS.QVloVoSEQpNssuLTNhQTKGRKlRRH+KEKEEKKRKKPVSQSlEFPTRYSPYRP
ZF12 (2114-2139) SMART E=0.44 zps=16.3 Canonical
YXCX4CX3FX5ψX2HX4H
1 1 1
■■■■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. YRCVHQGCFAAFTIQQNLILHYQAVH
100% YpCVHQGChAAFTIQQNLILHYQAVH
90% YpCVHQGCFAAFTIQQNLILHYQAVH
80% YRCVHQGCFAAFTIQQNLILHYQAVH
70% YRCVHQGCFAAFTIQQNLILHYQAVH
h+0h+p00hhhhphppphhh+hphh+
YCCVHQGCFAAFTIQQNLILHYQAVH (Mouse)
* ************************
YQCVHQGCTAAFTIQQNLILHYQAVH (Xenopus)
*:****** *****************
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Tyr2114del 1 5.77
p.Arg2115del 1 5.77
p.Cys2121Phe 1 0.999 206 1 5.54 21.7
p.Phe2122Ser 1 0.958 155 1 5.54 7.947
p.Ile2132Val 1 0.66 29 1 5.54 16.83
α=1 β=4
PWMs calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.309 1.000 0.010 0.222
c 0.313 0.000 0.001 0.157
g 0.309 0.000 0.086 0.000
t 0.069 0.000 0.903 0.620
Ent= 1.838 0.000 0.515 1.333
|
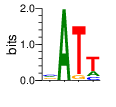
|
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.161 0.993 0.005 0.278
c 0.599 0.006 0.005 0.138
g 0.160 0.001 0.985 0.507
t 0.081 0.001 0.005 0.077
Ent= 1.582 0.070 0.138 1.689
|
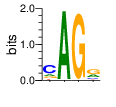
|
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.078 0.721 0.482
c 0.204 0.145 0.085
g 0.035 0.100 0.196
t 0.683 0.034 0.237
Ent= 1.299 1.241 1.763
|
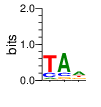
|
Linker 12-13 (2140-2171, 32aa)
■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■
h.s. KSDLPAFSAEVEEESEAGKESEETETKQTLKE
100% KSs...Fs.E.tE.........p.Est...+E
90% KSsLPsFSsElEEEsE.sK-p-EhEsK.shKE
80% KSDLPuFSAEVEEEsEssKEsEEhETKpohKE
70% KSDLPAFSAEVEEESEsGKESEEhETKQThKE
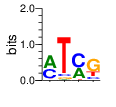
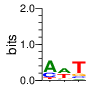
ZF13 (2172-2197) SMART E=.0472 zps=15 Canonical
FXCX4CX3FX5ψX2HX4H
1 13 1 12 11 1
■■■■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. FRCQVSDCSRIFQAITGLIQHYMKLH
100% FRC...sCSRIFQtlsuLlQHYMKhH
90% FRCphSDCSRIFQtlTuLIQHYMKLH
80% FRCQVSDCSRIFQAITGLIQHYMKLH
70% FRCQVSDCSRIFQAITGLIQHYMKLH
h+0php-0p+hhphhp0hhp+hh+h+
FRCQVSDCSRIFQAITGLIQHYMKLH (Mouse)
**************************
FRCMEIDCSRIFQEVGSLVQHYMKLH (Xenopus)
*** ******* : .*:*****:*
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Arg2173Gln 1 1 43 1 5.54 32
p.Ser2177Ile 1 0.974 142 1 5.54 18.61
p.Asp2178Asn 3 0.289 23 1 5.54 15.4
p.Ser2180Cys 1 1 112 1 5.54 22.9
p.Ala2185Ser 1 0.435 99 1 4.66 12.79
p.Ile2186Val 2 0.002 29 1 5.54 1.247
p.Ile2190Met 1 0.961 10 1 4.24 15.83
p.Gln2191Lys 1 0.993 53 1 5.42 17.59
p.Met2194Val 1 0.15 21 1 5.42 11.3
α=5 β=4
PWMs calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.251 0.000 0.963 0.001
c 0.251 0.000 0.028 0.016
g 0.250 0.000 0.004 0.980
t 0.247 1.000 0.005 0.002
Ent= 2.000 0.001 0.268 0.154
|
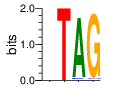
|
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.540 0.026 0.401 0.014
c 0.400 0.066 0.550 0.132
g 0.024 0.053 0.006 0.616
t 0.035 0.854 0.044 0.238
Ent= 1.310 0.816 1.246 1.396
|
|
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.599 0.438 0.083
c 0.261 0.073 0.114
g 0.024 0.101 0.155
t 0.116 0.388 0.648
Ent= 1.436 1.662 1.479
|
|
Linker 13-14 (2198-2215, 18aa)
■■■■■■■■■■■■■■■■■■
h.s. EMTPEEIESMTASVDVGK
100% pM.sEpItsh.s..phup
90% EMoPEEItSMpsulslG+
80% EMTPEEIESMTuuVDVGK
70% EMTPEEIESMTASVDVGK
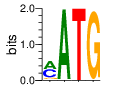
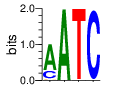
ZF14 (2216-2241) SMART E=0.498 zps=10.2 Canonical
FXCX4CX3FX5ψX2HX4H
9 11 1 1 2
■■■■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. FPCDQLECKSSFTTYLNYVVHLEADH
100% F.CDQ.pCt..FTshhsYl.HLE..H
90% FsCDQ.pCKSSFTsYLsYllHLEsDH
80% FPCDQ.ECKSSFTTYLNYVVHLEsDH
70% FPCDQLECKSSFTTYLNYVVHLEsDH
h00-ph-0+pphpphhphhh+h-h-+
FPCDQLECKLSFTTYLSYVVHLEVDH (Mouse)
********* ******.******.**
FKCDQLNCALLFTSCTSYIEHLEEVH (Xenopus)
* ****:* **: .*: *** *
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Lys2224Asn 9 0.999 94 0.998 -0.083 15.9
p.Thr2229Ala 1 0.002 58 0.984 3.75 8.2
p.Tyr2230His 1 0.989 83 0.978 4 18.4
p.Tyr2233Cys 1 1 194 1 5 17.78
p.Val2235Ile 1 0.002 29 0.989 -2.21 10.67
p.Asp2240Asn 1 0.115 23 1 5.63 15.2
p.Asp2240Glu 1 0 45 1 2.93 9.146
α=5 β=1
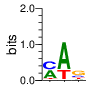
PWMs calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.444 0.997 0.000 0.000
c 0.442 0.000 0.000 0.010
g 0.098 0.003 0.001 0.990
t 0.016 0.000 0.999 0.000
Ent= 1.465 0.031 0.008 0.082
|
|
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.739 1.000 0.003 0.001
c 0.222 0.000 0.000 0.993
g 0.037 0.000 0.001 0.006
t 0.002 0.000 0.997 0.000
Ent= 0.999 0.006 0.034 0.066
|
|
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.440 0.728 0.089
c 0.450 0.005 0.202
g 0.025 0.011 0.520
t 0.086 0.256 0.189
Ent= 1.476 0.947 1.721
|
|
Linker 14-15 (2242-2255, 14aa)
■■■■■■■■■■■■■■
h.s. GIGLRASKTEEDGV
100% tht.+..+..t-th
90% ulth+ssKsE-DGh
80% GIGhRssKsE-DGl
70% GIGhRsSKTEEDGl
ZF15 (2256-2281) SMART E=0.164 zps=23 HMMER=2.2e-06 Canonical
YXCX4CX3YX5ψX2HX4H
1 11 2 1
■■■■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. YKCDCEGCDRIYATRSNLLRHIFNKH
100% aKCDCEGCDRlYATRSNLLRHIFNKH
90% aKCDCEGCDRIYATRSNLLRHIFNKH
80% YKCDCEGCDRIYATRSNLLRHIFNKH
70% YKCDCEGCDRIYATRSNLLRHIFNKH
h+0-0-00-+hhhp+pphh++hhp++
YKCDCEGCDRIYATRSNLLRHIFNKH (Mouse)
**************************
YKCDCEGCDRVYATRSNLLRHIFNKH (Xenopus)
**********:***************
gnomAD v2.1 Controls
Mutation N PolyPhen Grantham Phast GERP CADD
p.Lys2257Arg 1 0.997 26 1 5.65 15.51
p.Asp2264Gly 1 1 94 1 5.63 14.41
p.Arg2265Cys 1 1 181 1 4.68 15.89
p.Arg2270Trp 2 1 101 0.959 4.68 15.99
p.Asn2279Ser 1 0.997 46 1 5.62 14.92
α=2 β=3
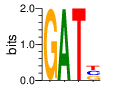
PWM calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.000 1.000 0.000 0.000
c 0.000 0.000 0.000 0.333
g 1.000 0.000 0.000 0.333
t 0.000 0.000 1.000 0.333
Ent= 0.000 0.000 0.000 1.587
|
|
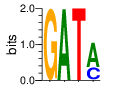
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.000 1.000 0.000 0.664
c 0.000 0.000 0.000 0.332
g 1.000 0.000 0.000 0.004
t 0.000 0.000 1.000 0.000
Ent= 0.000 0.000 0.001 0.954
|
|
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.260 0.576 0.221
c 0.281 0.116 0.245
g 0.385 0.154 0.250
t 0.074 0.154 0.284
Ent= 1.827 1.650 1.994
|

|
Linker 15-16 (2282-2385, 104)
■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■
h.s. NDKHKAHLIRPRRLTPGQENMSSKANQEKSKSKHRGTKHSRCGKEGIKMPKTKRKKKNNLENKNAKIVQIEENKPYSLKR
100% N-+HKtHLIRPR+.h.sQ-shspKs.p-+sh.+.......+.tpcu.c..+.+Rp................ppp.hoLKh
90% ND+HKsHLIRPR+LT.GQENhSSKANQEKshtK.Rsh...RsG+-G.+h.KsKRKKp.sh-sKs.Kh.php-sKsYSLKR
80% NDKHKAHLIRPR+LTsGQENhSSKANQEKsKsKaRGTKa.RsGKEGhKhsKTKRKKKsNLEsKsuKllQIEENKPYSLKR
70% NDKHKAHLIRPRRLTPGQENhSSKANQEKoKSKaRGTKH.RsGKEGhKhPKTKRKKKsNLENKsAKIVQIEENKPYSLKR
■■■■■■■■■■■■■■■■■■■■■■■■
h.s. GKHVYSIKARNDALSECTSRFVTQ
100% G+asa.lKs+ptAhsECssphhhQ
90% GKHVYSIKARN-ALSECTS+FVTQ
80% GKHVYSIKARNDALSECTS+FVTQ
70% GKHVYSIKARNDALSECTSRFVTQ
coiled-coil 2334-2354 (21)
ZF16 (2386-2410) SMART E=0.286 zps=7.6 Non-canonical (but V a smaller hyrophic aa replaces F at postion C2+3)
YXCX4CX9ψX2HX3H
1 2 2
■■■■■■■■■■■■■■■■■■■■■■■■■
-1123456789
h.s. YPCMIKGCTSVVTSESNIIRHYKCH
100% YPCMlpGCoSVVTSEpsIIRHYKCH
90% YPCMIKGCoSVVTSEsNIIRHYKCH
80% YPCMIKGCTSVVTSESNIIRHYKCH
70% YPCMIKGCTSVVTSESNIIRHYKCH
h00hh+00pphhpp-pphh++h+0+
YPCMIKGCTSVVTSESNIIRHYKCH (Mouse)
*************************
YPCMVRGCTSVVTSERSIIRHYKCH (Xenopus)
****::********* .********
gnomAD v2.1 Control
Mutation N PolyPhen Grantham Phast GERP CADD
p.Lys2391Met 1 1 95 1 5.63 13.75
p.Val2397Phe 2 0.999 5 1 5.43 15.36
p.Asn2402Ser 2 0.816 46 1 5.43 4.25
α=1 β=2
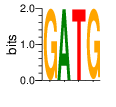
PWM calculated using zf.princeton.edu with Expanded Linear SVM
|
base 1 2 3 4
a 0.000 1.000 0.000 0.000
c 0.000 0.000 0.000 0.000
g 1.000 0.000 0.002 0.998
t 0.000 0.000 0.998 0.002
Ent= 0.000 0.000 0.022 0.023
|
|
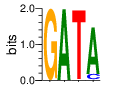
PWM calculated using zf.princeton.edu with Polynomial SVM
|
base 1 2 3 4
a 0.000 1.000 0.002 0.888
c 0.000 0.000 0.002 0.112
g 1.000 0.000 0.003 0.001
t 0.000 0.000 0.993 0.000
Ent= 0.000 0.000 0.070 0.513
|
|
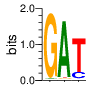
PWM calculated using zf.princeton.edu with B1H RFR
|
base 1 2 3
a 0.016 0.953 0.007
c 0.001 0.008 0.166
g 0.981 0.019 0.018
t 0.002 0.020 0.809
Ent= 0.149 0.341 0.833
|
|
coiled-coil 2530-2550
LKRVNKEKNVSQNKKRKVEK
|
