 |
HOME CONSERVATION C-TERMINAL ANALYSES DNA AND RNA SEQUENCES RESEARCH QUESTIONS RESULTS RESOURCES REFERENCES |
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Though the zinc finger containing regions of ZNF292, RLF and ZNF654 are divergent, their N-terminal domains (here defined as the sequence up to first zinc finger) are highly similar. The greatest similarity is between ZNF292 and RLF (51.71%). C2H2 Zinc Fingers are often coupled with N-terminal domains that contain well known domains such as KRAB, SCAN or POZ/BTB. None of the commonly used online sequence analysis tools identify any of these domains in these three proteins. The homologous region in ZNF292 is composed of ~24 alpha helices. Many of them are amphipathic with a well-defined hydrophobic face which is indicative of one or more globular regions. Protein (range) Protein (range) Similarity Identities Positives Gaps E-Value ZNF292 (29:540) to RLF (53:553) 51.73% 52% 69% 5% 2e-179 ZNF292 (46:466) to ZNF654 (57:471) 37.83% 38% 57% 1% 5e-100 ZNF654 (51:470) to RLF (68:488) 38.68% 39% 56% 1% 4e-92 ZNF292 N-Terminal Secondary Structure Prediction ZNF292 N-terminal secondary structure prediction details 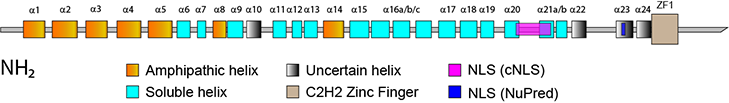
RLF N-Terminal Secondary Structure Prediction RLF N-terminal secondary structure prediction details 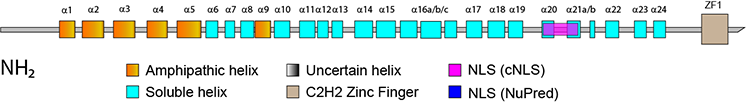
ZNF654 N-Terminal Secondary Structure Prediction ZNF654 N-terminal secondary structure prediction details 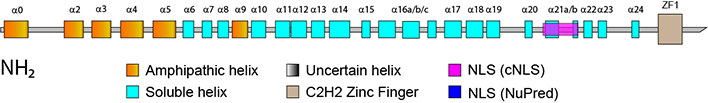
Characteristic Values of Predicted Secondary Structures Comparison of hydrophobicity, hydrophobic moment and charge of corresponding predicted alpha-helices in the N-terminal region of ZNF292 and paralogs RLF and ZNF654.
Alignments with Secondary Structure Predictions ZNF292-RLF-ZNF654 Alignment with Consensus Secondary Structure Predictions ZNF292 MADEEAE--------------------------------QERLSCGEGGCV--AELQRLG 26 hhhHHHH HHHhcccccchH HHHHHHH RLF MADGKGDAAAVAGAGAEAP----------AVAGAGDGVETESMVRGHRPVSPAPGASGLR 50 chcchhhHHHHhhccccch hhhcccchhhHHHHhhchccccchhHhHhHH ZNF654 MAEEESDQEAER-LGEELVAIVESPLGPVGLRAAGDGRG----GAGSGNCGG-------- 47 hhhhhHHHHHHH HHHHHHHHHHhhhhhhhhhhhhhhhhh hhhhcccc MA: :.: G α0 α1 ZNF292 ERLQELELQLRESRVPAVEAATDYCQQLCQTLLEYAEKWKTSEDPLPLLEVYTVAIQSYV 86 HHHHHHHHHHHHhhchHHHHHHHHHHHHHHHHHHHHHHhcccccchHHHHHHHHHHHHHH RLF PCLWQLETELREQEVSEV-SSLNYCRSFCQTLLQYASNKNASEHIVYLLEVYRLAIQSFA 109 HHHHHHHHHHHhhhhhHH HHHHHHHHHHHHHHHHHHhcchhhhHHHHHHHHHHHHHHHH ZNF654 ---------------GVGISSRDYCRRFCQVVEDYAGRWQV---PLPQLQVLQTALCCFT 89 chhhhHHHHHHHHHHHHHHHHchccc chHHHHHHHHHHHHHH :: :YC: :CQ.: :YA . :. : L:V A: .:. α2 α3 ZNF292 KARPYLTSECENVALVLERLALSCVELLLCLP-VELSDKQWEQFQTLVQVAHEKLMENGS 145 HhhhhcchhHHHHHHHHHHHHHHHHHHHHhcc ccccHHHHHHHHHHHHHHHHHHHHhcc RLF SARPYLTTECEDVLLVLGRLVLSCFELLLSVSESELPCEVWLPFLQSLQESHDALLEFGN 169 HhhhccccchHHHHHHHHHHHHHHHHHHhhcccccccHHHHHHHHHHHHHHHHHHHHhcc ZNF654 TASASFPDECEHVQYVLSSLAVSFFELLLFFGRDEFYEEPLKDILGSFQECQNHLRRYGN 149 HhhhhcccchHHHHHHHHHHHHHHHHHHHHcchhhhhHHHHHHHHHHHHHHHHHHHHhcc .A : ECE.V VL L.:S .ELLL . E: : : .Q .:: L . G. α4 α5 ZNF292 CELHFLATLAQETGVWKNPVLCTILSQEPLDKDKVNEFLAFEGPILLDMRIKHLIKTNQL 205 hHHHHHHHHHHhccccccHHHHHHHccccccHHHHHHHHHHhhhHHHHHHHHHHHHHhhH RLF NNLQILVHV-TKEGVWKNPVLLKILSQQPVETEEVNKLIAQEGPSFLQMRIKHLLKSNCI 228 hhHHHHHH HHhhcccccHHHHHHHhcccccHHHHHHHHHHhchHHHHHHHHHHHHhcch ZNF654 VNLELVTRIIRDGGPWEDPVLQAVLKAQPASQEIVNKYLSSENPLFFELRARYLIACERI 209 hhHHHHHHHHHcccccchHHHHHHHHhccccHHHHHHHHHhcchHHHHHHHHHHHHHchh :L.::. : . G W::PVL :L. :P . : VN: :: E.P ::::R ::L: : : α6 α7 α8 α9 ZNF292 SQATALAKLCSDHPEIGIKGSFKQTYLVCLCTSSPNGKLI-EEISEVDCKDALEMICNLE 264 HHHHHHHHHHhhchhhcchhHHhHhhhhhhhhccchHHHH HHHHHhhHHHHHHHHHHHh RLF PQATALSKLCAESKEISNVSSFQQAYITCLCSMLPNEDAI-KEIAKVDCKEVLDIICNLE 287 HHHHHHHHHHHhchhhcchhHHHHHHHHHHHHhcchHHHH HHHHHhhHHHHHHHHHHHh ZNF654 PEAMALIKSCINHPEISKDLYFHQALFTCLFMSPVEDQLFREHLLKTDCKSGIDIICNAE 269 HHHHHHHHHHhhchhhchhHHHHHHHHHHHHHhhhHHHHHHHHHHHcchHHHHHHHHHhh :A AL K C : EI. F:Q: :.CL : . : :.: :.DCK. :::ICN E α10 α11 α12 α13 ZNF292 SEGDEKSALVLCTAFLSRQLQQGDMYCAWELTLFWSKLQQRVEPSIQVYLERCRQLSLLT 324 hcchhHHHhhhHHHHHHHHHHccchHHHHHHHHHHHHHHHHhcchHHHHHHHHHHHHHHH RLF SEGQDNTAFVLCTTYLTQQLQTASVYCSWELTLFWSKLQRRIDPSLDTFLERCRQFGVIA 347 hccchhHhhhhhHHHHHHHHhhcchhHHHHHHHHHHHHHHHccchHHHHHHHHHHHHHHH ZNF654 KEGKTMLALQLCESFLIPQLQNGDMYCIWELIFIWSKLQLKSNPSKQVFVDQCYQLLRTA 329 hcccHHHHHHHHHHHHHHHHHccchhhhhHHHHHHHHHhhhccccHHHHHHHHHHHHHHh .EG. A: LC ::L QLQ ..:YC WEL ::WSKLQ : :PS :.::::C Q: : α14 α15 α16a ZNF292 KTVYHIFFLIKVINSETEGAGLATCIELCVKALRLESTENTEVKISICKTISCLLPDDLE 384 HHHHHHHHHHHHhhhhhhhhhHHHHHHHHHHHHHhccccchhHHHHHHHHHHHHhhchHH RLF KTQQHLFCLIRVIQTEAQDAGLGVSILLCVRALQLRSSEDEEMKASVCKTIACLLPEDLE 407 HHHHHHHHHHHHHHHHHHHcchHHHHHHHHHHHhccccccHHHHHHHHHHHHHHcccHHH ZNF654 TNVRVIFPFMKIIKDEVEEEGLQICVEICGCALQLDLHDDPKTKCLIYKTIAHFLPNDLE 389 hhHhhhhHHHHHHHHHHHHHhHHHHHHHhhhhhhcccccchhHHHHHHHHHHHHccccHH .. :F ::::I: E.: GL .: :C AL:L :: : K : KTI: :LP:DLE α16b α16c α17 ZNF292 VKRACQLSEFLIEPTVDAYYAVEMLYNQPDQKYDEENLPIPNSLRCELLLVLKTQWPFDP 444 HHHHHHHHHHHHcccHHHHHHHHHHHhcccccccccccccchhhhhHHHhhHHccccccc RLF VRRACQLTEFLIEPSLDGFNMLEELYLQPDQKFDEENAPVPNSLRCELLLALKAHWPFDP 467 HHHHHHHHHHHHcccHHHHHHHHHHHhcccccccccccccchhhHHHHHHHHHhccccch ZNF654 ILRICALSIFFLERSLEAYRTVEELYKRPDEEYNEGTSSVQNRVRFELLPILKKGLFFDP 449 HHHHHHHHHHHHhhhHHHHHHHHHHHhccccccccccchhhhhhhhHHHHHHHhccccch : R C L: F::E :::.: :E LY :PD::::E . : N :R ELL LK FDP α18 α19 α20 ZNF292 EFWDWKTLKRQCLALMGEEASIVSSI-----D---ELNDSEVY----EKVVDYQEESKE- 491 chhhHHHHHHHHHHHHhhhhhhhhhh h hcchhhhh hhhhhhhhhccc RLF EFWDWKTLKRHCHQLLGQEASDSDDDLSGYEM---SINDTDVL----ESFLSDYD----- 515 chhhHHHHHHHHHHHHhhhhhhhhhhhhhhhc chchhhhh hhHhhhcc ZNF654 EFWNFVMIKKNCVALLSDKSAVRFLNESTLENNAGNLKRTEEQQGLDEGFDSLTDQSTGE 509 hHHhHHHHHHHHHHHHhhhHHhhhhhHHhHhhhhhcccchhhhhchhhhhhhhccccccc EFW:: :K::C L:.:::: .:: :: E . . : α21a α21b α22 ZNF292 TSMNGLSGGVGANS--GLLKDIGDEKQKKREIKQLRERGFISARFRNWQAYMQY 543 ccccccccccchhh hhhhhhhhHHHHHHHHHhhhhccchhHHHhhHHHHHhh RLF -------EGKEDKQ--YRRRDLTDQHKEKRDKKPI----GSSERYQRWLQYKFF 556 ccccccc hhHcchhhHHHHHhccccc cchHHHHHHHHhhhh ZNF654 TDPDDV-SGVQPKGHINTKKNLTALSTSKVDHNVPRHR---------------- 546 cccccc cccccccccccccccchhcccccccccccch G : ::: .K : : α23 α24 ZNF292-RLF Alignment 1 36 hh---h -p-+hpss-sssh h-hp+hs-+hp-h-hph hhhHHH HHHHhcccccchH HHHHHHHHHHHHHHHHH cons ■■■■■■----------------------■■■■■■■■■■■■■--■■■■■■■■■■■■■■■■■ ZNF292 MADEEA----------------------EQERLSCGEGGCV--AELQRLGERLQELELQL MAD :. E E : G. . L L :LE :L RLF MADGKGDAAAVAGAGAEAPAVAGAGDGVETESMVRGHRPVSPAPGASGLRPCLWQLETEL cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ chcchhhHHHHhhccccchhhhcccchhhHHHHhhchccccchhHhHhHHHHHHHHHHHH hh-s+s-hhhhhshsh-hshhhshs-sh-p-phh+s++shpshsshpsh+sshhph-p-h 1 60 α0 α1 37 96 +-p+hshh-hhp-hspphspphh-hh-+h+pp--shshh-hhphhhpphh+h+shhpp-s HHhhchHHHHHHHHHHHHHHHHHHHHHHhcccccchHHHHHHHHHHHHHHHhhhhcchhH cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ ZNF292 RESRVPAVEAATDYCQQLCQTLLEYAEKWKTSEDPLPLLEVYTVAIQSYVKARPYLTSEC RE..V. V.: . YC:.:CQTLL:YA.: ::SE. : LLEVY :AIQS:..ARPYLT:EC RLF REQEVSEV-SSLNYCRSFCQTLLQYASNKNASEHIVYLLEVYRLAIQSFASARPYLTTEC cons ■■■■■■■■-■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ HhhhhhHH HHHHHHHHHHHHHHHHHHhcchhhhHHHHHHHHHHHHHHHHHhhhccccch +-p-hp-h pphphs+phspphhphhpp+php-+hhhhh-hh+hhhpphhph+shhpp-s 61 119 α2 α3 97 155 -phhhhh-+hhhpsh-hhhshs h-hp-+ph-phpphhphh+-+hh-psps-h+hhhphh HHHHHHHHHHHHHHHHHHHhcc ccccHHHHHHHHHHHHHHHHHHHHhcchHHHHHHHHH cons ■■■■■■■■■■■■■■■■■■■■■■-■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ ZNF292 ENVALVLERLALSCVELLLCLP-VELSDKQWEQFQTLVQVAHEKLMENGSCELHFLATLA E:V LVL RL.LSC.ELLL.:. EL. : W F :Q :H: L:E G. :L::L. : RLF EDVLLVLGRLVLSCFELLLSVSESELPCEVWLPFLQSLQESHDALLEFGNNNLQILVHV- cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■- HHHHHHHHHHHHHHHHHHhhcccccccHHHHHHHHHHHHHHHHHHHHhcchhHHHHHHH --hhhhhs+hhhpsh-hhhphp-p-hss-hhhshhpphp-p+-hhh-hsppphphhh+h 120 178 α4 α5 α6 156 215 p-pshh+pshhsphhpp-sh-+-+hp-hhhh-sshhh-h+h++hh+ppphpphphhh+hs HhccccccHHHHHHHccccccHHHHHHHHHHhhhHHHHHHHHHHHHHhhHHHHHHHHHHH cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ ZNF292 QETGVWKNPVLCTILSQEPLDKDKVNEFLAFEGPILLDMRIKHLIKTNQLSQATALAKLC :E VWKNPVL .ILSQ:P::.::VN:::A EGP :L:MRIKHL:K:N :.QATAL:KLC RLF TKEGVWKNPVLLKILSQQPVETEEVNKLIAQEGPSFLQMRIKHLLKSNCIPQATALSKLC cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ hhhcccccHHHHHHHhcccccHHHHHHHHHHhchHHHHHHHHHHHHhcchHHHHHHHHHH p+-shh+pshhh+hhpppsh-p--hp+hhhp-ssphhph+h++hh+ppshsphphhp+hs 180 238 α7 α8 α9 α10 216 275 p-+s-hsh+sph+pphhhshspppsps+hh--hp-h-s+-hh-hhsph-p-s--+phhhh hhchhhcchhHHhHhhhhhhhhccchHHHHHHHHHhhHHHHHHHHHHHhhcchhHHHhhh cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ ZNF292 SDHPEIGIKGSFKQTYLVCLCTSSPNGKLIEEISEVDCKDALEMICNLESEGDEKSALVL :: EI. .SF:Q:Y:.CLC: PN . I:EI::VDCK:.L::ICNLESEG::::A:VL RLF AESKEISNVSSFQQAYITCLCSMLPNEDAIKEIAKVDCKEVLDIICNLESEGQDNTAFVL cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ HhchhhcchhHHHHHHHHHHHHhcchHHHHHHHHHhhHHHHHHHHHHHhhccchhHhhhh h-p+-hpphpphpphhhpshsphhsp--hh+-hh+h-s+-hh-hhsph-p-sp-pphhhh 239 298 α11 α12 α13 α14 276 335 sphhhp+phpps-hhshh-hphhhp+hpp+h-sphphhh-+s+phphhp+phh+hhhhh+ HHHHHHHHHHccchHHHHHHHHHHHHHHHHhcchHHHHHHHHHHHHHHHHHHHHHHHHHH cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ ZNF292 CTAFLSRQLQQGDMYCAWELTLFWSKLQQRVEPSIQVYLERCRQLSLLTKTVYHIFFLIK CT::L::QLQ ..:YC:WELTLFWSKLQ:R::PS::.:LERCRQ:.:::KT H:F LI: RLF CTTYLTQQLQTASVYCSWELTLFWSKLQRRIDPSLDTFLERCRQFGVIAKTQQHLFCLIR cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ hHHHHHHHHhhcchhHHHHHHHHHHHHHHHccchHHHHHHHHHHHHHHHHHHHHHHHHHH spphhppphpphphhsph-hphhhp+hp++h-sph-phh-+s+phshhh+ppp+hhshh+ 299 358 α15 α16a 336 395 hhpp-p-shshhpsh-hsh+hh+h-pp-pp-h+hphs+phpshhs--h-h++hsphp-hh HhhhhhhhhhHHHHHHHHHHHHHhccccchhHHHHHHHHHHHHhhchHHHHHHHHHHHHH cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ ZNF292 VINSETEGAGLATCIELCVKALRLESTENTEVKISICKTISCLLPDDLEVKRACQLSEFL VI::E::.AGL...I LCV:AL:L.S:E: E:K S:CKTI:CLLP:DLEV:RACQL:EFL RLF VIQTEAQDAGLGVSILLCVRALQLRSSEDEEMKASVCKTIACLLPEDLEVRRACQLTEFL cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ HHHHHHHHcchHHHHHHHHHHHhccccccHHHHHHHHHHHHHHcccHHHHHHHHHHHHHH hhpp-hp-hshshphhhsh+hhph+pp----h+hphs+phhshhs--h-h++hsphp-hh 359 418 α16b α16c α17 α18 396 455 h-sph-hhhhh-hhhpps-p+h---phshspph+s-hhhhh+pphsh-s-hh-h+ph++p HcccHHHHHHHHHHHhcccccccccccccchhhhhHHHhhHHcccccccchhhHHHHHHH cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ ZNF292 IEPTVDAYYAVEMLYNQPDQKYDEENLPIPNSLRCELLLVLKTQWPFDPEFWDWKTLKRQ IEP::D.: :E LY QPDQK:DEEN P:PNSLRCELLL.LK::WPFDPEFWDWKTLKR: RLF IEPSLDGFNMLEELYLQPDQKFDEENAPVPNSLRCELLLALKAHWPFDPEFWDWKTLKRH cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ HcccHHHHHHHHHHHhcccccccccccccchhhHHHHHHHHHhccccchchhhHHHHHHH h-sph-shphh--hhhps-p+h---phshspph+s-hhhhh+h+hsh-s-hh-h+ph+++ 419 478 α19 α20 α21 456 510 shhhhs--hphhpph --hp-p-hh-+hh-hp--p+-pphpshpsshshppshh+- HHHHHhhhhhhhhhh hhcchhhhhhhhhhhhhhcccccccccccccchhhhhhhh cons ■■■■■■■■■■■■■■■-----■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ ZNF292 CLALMGEEASIVSSI-----DELNDSEVYEKVVDYQEESKETSMNGLSGGVGANSGLLKD C L:G:EAS .. .:LD::V E..:. . G :. :D RLF CHQLLGQEASDSDDDLSGYEMSINDTDVLESFLSDY-----------DEGKEDKQYRRRD cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■-----------■■■■■■■■■■■■■ HHHHHhhhhhhhhhhhhhhhcchchhhhhhhHhhhc cccccccchhHcc s+phhsp-hp-p---hpsh-hphp-p-hh-phhp-h --s+--+ph+++- 479 527 α22 516 548 hs--+p+++-h+ph+-+shhph+h+phphhhsh hhhHHHHHHHHHhhhhccchhHHHhhHHHHHhh cons ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ ZNF292 IGDEKQKKREIKQLRERGFISARFRNWQAYMQY : D::::KR: K S R::.W Y : RLF LTDQHKEKRDKKP----IGSSERYQRWLQYKFF cons ■■■■■■■■■■■■■----■■■■■■■■■■■■■■■■ hhhHHHHHhcccc ccchHHHHHHHHhhhh hp-p++-++-++s hspp-+hp+hhph+hh 533 561 α23 α24 ZNF292-ZNF654 Alignment 1 54 hh---h-p-+hpss-ssshh-hp+hs-+hp-h-hph+ -p+hshh-hhp-hspph hhhHHHHHHHhcccccchHHHHHHHHHHHHHHHHHHH HhhchHHHHHHHHHHHH ZNF292 MADEEAEQERLSCGEGGCVAELQRLGERLQELELQLR------ESRVPAVEAATDYCQQL MA:EE::QE GE . . LG . : R :. . :: DYC::: ZNF654 MAEEESDQEAERLGEELVAIVESPLGPVGLRAAGDGRGGAGSGNCGGGVGISSRDYCRRF hhhhhHHHHHHHHHHHHHHHHHhhhhhhhhhhhhhhhhhhhhhccccchhhhHHHHHHHH hh---p-p-h-+hs--hhhhh-pshsshsh+hhs-s+sshspspsssshshpp+-hs++h 1 60 α0 α1 α2 55 114 spphh-hh-+h+pp--shshh-hhphhhpphh+h+shhpp-s-phhhhh-+hhhpsh-hh HHHHHHHHHHhcccccchHHHHHHHHHHHHHHHhhhhcchhHHHHHHHHHHHHHHHHHHH ZNF292 CQTLLEYAEVWKTSEDPLPLLEVYTVAIQSYVKARPYLTSECENVALVLERLALSCVELL MA.: :YA :W:.. LP L:V .A: .:..A . :..ECE:V VL. LA:S .ELL ZNF654 CQVVEDYAGRWQVP---LPQLQVLQTALCCFTTASASFPDECEHVQYVLSSLAVSFFELL HHHHHHHHchcccc hHHHHHHHHHHHHHHHhhhhcccchHHHHHHHHHHHHHHHHHH sphh--hhs+hphs hsphphhpphhsshpphphphs--s-+hphhhpphhhphh-hh 61 117 α3 α4 115 173 hshs h-hp-+ph-phpphhphh+-+hh-psps-h+hhhphhp-pshh+pshhsphhpp- Hhcc ccccHHHHHHHHHHHHHHHHHHHHhcchHHHHHHHHHHhccccccHHHHHHHccc ZNF292 LCLP-VELSDKQWEQFQTLVQVAHEKLMENGSCELHFLATLAQETGVWKNPVLCTILSQE L : E: :: ::: .Q .:::L . G. :L.::: : :: G W::PVL ::L. : ZNF654 LFFGRDEFYEEPLKDILGSFQECQNHLRRYGNVNLELVTRIIRDGGPWEDPVLQAVLKAQ HHcchhhhhHHHHHHHHHHHHHHHHHHHHhcchhHHHHHHHHHcccccchHHHHHHHHhc hhhs+--hh--sh+-hhsphp-spp+h++hsphph-hhp+hh+-sssh--shhphhh+hp 118 177 α5 α6 α7 174 233 sh-+-+hp-hhhh-sshhh-h+h++hh+ppphpphphhh+hsp-+s-hsh+sph+pphhh cccHHHHHHHHHHhhhHHHHHHHHHHHHHhhHHHHHHHHHHHhhchhhcchhHHhHhhhh ZNF292 PLDKDKVNEFLAFEGPILLDMRIKHLIKTNQLSQATALAKLCSDHPEIGIKGSFKQTYLV P .:: VN::L: E.P:::::R ::LI :::.:A AL K C :HPEI. . F:Q: :. ZNF654 PASQEIVNKYLSSENPLFFELRARYLIACERIPEAMALIKSCINHPEISKDLYFHQALFT cccHHHHHHHHHhcchHHHHHHHHHHHHHchhHHHHHHHHHHhhchhhchhHHHHHHHHH shpp-hhp+hhpp-pshhh-h+h+hhhhs-+hs-hhhhh+pshp+s-hp+-hhh+phhhp 178 237 α8 α9 α10 α11 234 292 shspp psps+hh--hp-h-s+-hh-hhsph-p-s--+phhhhsphhhp+phpps-hhsh hhhhc cchHHHHHHHHHhhHHHHHHHHHHHhhcchhHHHhhhHHHHHHHHHHccchHHH ZNF292 CLCTS-SPNGKLIEEISEVDCKDALEMICNLESEGDEKSALVLCTAFLSRQLQYQGDMCA CL * : : E.: :.DCK..:::ICN E.EG. AL LC :FL QLQ:GDMYCA ZNF654 CLFMSPVEDQLFREHLLKTDCKSGIDIICNAEKEGKTMLALQLCESFLIPQLQNGDMYCI HHHHhhhHHHHHHHHHHHcchHHHHHHHHHhhhcccHHHHHHHHHHHHHHHHHccchhhh shhhpsh--phh+-+hh+p-s+psh-hhsph-+-s+phhhhphs-phhhsphpps-hhsh 238 297 α12 α13 α14 293 352 h-hphhhp+hpp+h-sphphhh-+s+phphhp+phh+hhhhh+hhpp-p-shshhpsh-h HHHHHHHHHHHHHhcchHHHHHHHHHHHHHHHHHHHHHHHHHHHhhhhhhhhhHHHHHHH ZNF292 WELTLFWSKLQQRVEPSIQVYLERCRQLSLLTKTVYHIFFLIKVINSETEGAGLATCIEL WEL ::WSKLQ : :PS QV::::C QL :..V IF ::K:I:.E.E GL C:E: ZNF654 WELIFIWSKLQLKSNPSKQVFVDQCYQLLRTATNVRVIFPFMKIIKDEVEEEGLQICVEI hHHHHHHHHHhhhccccHHHHHHHHHHHHHHhhhHhhhhHHHHHHHHHHHHHhHHHHHHH h-hhhhhp+hph+ppsp+phhh-pshphh+phpph+hhhshh+hh+--h---shphsh-h 298 357 α15 α16a α16b α16c 353 412 sh+hh+h-pp-pp-h+hphs+phpshhs--h-h++hsphp-hhh-sph-hhhhh-hhhpp HHHHHHhccccchhHHHHHHHHHHHHhhchHHHHHHHHHHHHHHcccHHHHHHHHHHHhc ZNF292 CVKALRLESTENTEVKISICKTISCLLPDDLEVKRACQLSEFLIEPTVDAYYAVEMLYNQ C AL:L: ::.:.K I KTI: :LP:DLE: R C LS F::E :::AY :VE LY:: ZNF654 CGCALQLDLHDDPKTKCLIYKTIAHFLPNDLEILRICALSIFFLERSLEAYRTVEELYKR hhhhhhcccccchhHHHHHHHHHHHHccccHHHHHHHHHHHHHHhhhHHHHHHHHHHHhc ssshhph-h+--s+p+shhh+phh+hhsp-h-hh+hshhphhhh-+ph-hh+ph--hh++ 358 417 α17 α18 α19 413 472 s-p+h---phshspph+s-hhhhh+pphsh-s-hh-h+ph++pshhhhs--hphhpph-- ccccccccccccchhhhhHHHhhHHcccccccchhhHHHHHHHHHHHHhhhhhhhhhhhh ZNF292 PDQKYDEENLPIPNSLRCELLLVLKTQWPFDPEFWDWKTLKRQCLALMGEEASIVSSIDE PD::Y:E . .: N :R ELL :LK. FDPEFW:: :K::C:AL:.::::: :. ZNF654 PDEEYNEGTSSVQNRVRFELLPILKKGLFFDPEFWNFVMIKKNCVALLSDKSAVRFLNES cccccccccchhhhhhhhHHHHHHHhccccchhHHhHHHHHHHHHHHHhhhHHhhhhhHH s---hp-sppphpp+h+h-hhshh++shhh-s-hhphhhh++pshhhhp-+phh+hhp-p 418 477 α20 α21 473 532 hp-p-hh-+hh-hp--p+-pphpshpsshshppshh+-hs--+p+++-h+ph+-+shhph cchhhhhhhhhhhhhhcccccccccccccchhhhhhhhhhhHHHHHHHHHhhhhccchhH ZNF292 LNDSEVYEKVVDYQEESKETSMNGLSGGVGANSGLLKDIGDEKQKKREIKQLRERGFISA :.:. : :::. : .::.L:. .::. . G Q K .I: :: :S: ZNF654 TLENNAGNLKRTEEQQGLDEGFDSLTDQSTGETDPDDVSG--VQPKGHINTKKNLTALST hHhhhhhcccchhhhhchhhhhhhhccccccccccccccc cccccccccccccchhcc ph-pphsph++p--ppsh--sh-php-ppps-p-s--hps hps+s+hpp++phphhpp 478 535 α22 α23 533 543 +h+phphhhsh HHhhHHHHHhh ZNF292 RFRNWQAYMQY : :. ZNF654 SKVDHNVPRHR cccccccccch p+h-+phs+++ 536 544 α24 N-terminal Content Analysis ZNF292 (29:540) LQELELQLRESRVPAVEAATDYCQQLCQTLLEYAEKWKTSEDPLPLLEVYTVAIQSYVKA RPYLTSECENVALVLERLALSCVELLLCLPVELSDKQWEQFQTLVQVAHEKLMENGSCEL HFLATLAQETGVWKNPVLCTILSQEPLDKDKVNEFLAFEGPILLDMRIKHLIKTNQLSQA TALAKLCSDHPEIGIKGSFKQTYLVCLCTSSPNGKLIEEISEVDCKDALEMICNLESEGD EKSALVLCTAFLSRQLQQGDMYCAWELTLFWSKLQQRVEPSIQVYLERCRQLSLLTKTVY HIFFLIKVINSETEGAGLATCIELCVKALRLESTENTEVKISICKTISCLLPDDLEVKRA CQLSEFLIEPTVDAYYAVEMLYNQPDQKYDEENLPIPNSLRCELLLVLKTQWPFDPEFWD WKTLKRQCLALMGEEASIVSSIDELNDSEVYEKVVDYQEESKETSMNGLSGGVGANSGLL KDIGDEKQKKREIKQLRERGFISARFRNWQAY UniProt % ZNF292 N % ZNF292/UniProt Ala (A) 9.18 Ala (A) 30 5.9 0.6427 Arg (R) 5.75 Arg (R) 18 3.5 0.6087 Asn (N) 3.84 Asn (N) 16 3.1 0.8073 Asp (D) 5.48 Asp (D) 23 4.5 0.8212 Cys (C) 1.19 Cys (C) 22 4.3 3.6134 Gln (Q) 3.77 Gln (Q) 31 6.1 1.6180 Glu (E) 6.17 Glu (E) 57 11.1 1.7990 Gly (G) 7.33 Gly (G) 18 3.5 0.4775 His (H) 2.18 His (H) 5 1 0.4587 Ile (I) 5.69 Ile (I) 25 4.9 0.8612 Leu (L) 9.9 Leu (L) 76 14.8 1.4949 Lys (K) 4.93 Lys (K) 34 6.6 1.3387 Met (M) 2.38 Met (M) 7 1.4 0.5882 Phe (F) 3.92 Phe (F) 14 2.7 0.6888 Pro (P) 4.85 Pro (P) 18 3.5 0.7216 Ser (S) 6.64 Ser (S) 35 6.8 1.0241 Thr (T) 5.55 Thr (T) 26 5.1 0.9189 Trp (W) 1.3 Trp (W) 9 1.8 1.3846 Tyr (Y) 2.92 Tyr (Y) 16 3.1 1.0616 Val (V) 6.91 Val (V) 32 6.2 0.8973 RLF (53:553) LWQLETELREQEVSEVSSLNYCRSFCQTLLQYASNKNASEHIVYLLEVYRLAIQSFASAR PYLTTECEDVLLVLGRLVLSCFELLLSVSESELPCEVWLPFLQSLQESHDALLEFGNNNL QILVHVTKEGVWKNPVLLKILSQQPVETEEVNKLIAQEGPSFLQMRIKHLLKSNCIPQAT ALSKLCAESKEISNVSSFQQAYITCLCSMLPNEDAIKEIAKVDCKEVLDIICNLESEGQD NTAFVLCTTYLTQQLQTASVYCSWELTLFWSKLQRRIDPSLDTFLERCRQFGVIAKTQQH LFCLIRVIQTEAQDAGLGVSILLCVRALQLRSSEDEEMKASVCKTIACLLPEDLEVRRAC QLTEFLIEPSLDGFNMLEELYLQPDQKFDEENAPVPNSLRCELLLALKAHWPFDPEFWDW KTLKRHCHQLLGQEASDSDDDLSGYEMSINDTDVLESFLSDYDEGKEDKQYRRRDLTDQH KEKRDKKPIGSSERYQRWLQY UniProt % RLF N % RLF/UniProt Ala (A) 9.18 Ala (A) 26 5.2 0.5664 Arg (R) 5.75 Arg (R) 23 4.6 0.8000 Asn (N) 3.84 Asn (N) 17 3.4 0.8854 Asp (D) 5.48 Asp (D) 28 5.6 1.0219 Cys (C) 1.19 Cys (C) 21 4.2 3.5294 Gln (Q) 3.77 Gln (Q) 35 7 1.8568 Glu (E) 6.17 Glu (E) 50 10 1.6207 Gly (G) 7.33 Gly (G) 13 2.6 0.3547 His (H) 2.18 His (H) 9 1.8 0.8257 Ile (I) 5.69 Ile (I) 22 4.4 0.7733 Leu (L) 9.9 Leu (L) 76 15.2 1.5354 Lys (K) 4.93 Lys (K) 26 5.2 1.0548 Met (M) 2.38 Met (M) 5 1 0.4202 Phe (F) 3.92 Phe (F) 18 3.6 0.9184 Pro (P) 4.85 Pro (P) 17 3.4 0.7010 Ser (S) 6.64 Ser (S) 42 8.4 1.2651 Thr (T) 5.55 Thr (T) 22 4.4 0.7928 Trp (W) 1.3 Trp (W) 9 1.8 1.3846 Tyr (Y) 2.92 Tyr (Y) 14 2.8 0.9589 Val (V) 6.91 Val (V) 28 5.6 0.8104 ZNF654 (52:469) SSRDYCRRFCQVVEDYAGRWQVPLPQLQVLQTALCCFTTASASFPDECEHVQYVLSSLAV SFFELLLFFGRDEFYEEPLKDILGSFQECQNHLRRYGNVNLELVTRIIRDGGPWEDPVLQ AVLKAQPASQEIVNKYLSSENPLFFELRARYLIACERIPEAMALIKSCINHPEISKDLYF HQALFTCLFMSPVEDQLFREHLLKTDCKSGIDIICNAEKEGKTMLALQLCESFLIPQLQN GDMYCIWELIFIWSKLQLKSNPSKQVFVDQCYQLLRTATNVRVIFPFMKIIKDEVEEEGL QICVEICGCALQLDLHDDPKTKCLIYKTIAHFLPNDLEILRICALSIFFLERSLEAYRTV EELYKRPDEEYNEGTSSVQNRVRFELLPILKKGLFFDPEFWNFVMIKKNCVALLSDKSAV UniProt % ZNF654 N % ZNF654/UniProt Ala (A) 9.18 Ala (A) 22 5.2 0.5664 Arg (R) 5.75 Arg (R) 21 5 0.8696 Asn (N) 3.84 Asn (N) 15 3.6 0.9375 Asp (D) 5.48 Asp (D) 21 5 0.9124 Cys (C) 1.19 Cys (C) 20 4.8 4.0336 Gln (Q) 3.77 Gln (Q) 23 5.5 1.4589 Glu (E) 6.17 Glu (E) 37 8.8 1.4263 Gly (G) 7.33 Gly (G) 13 3.1 0.4229 His (H) 2.18 His (H) 7 1.7 0.7798 Ile (I) 5.69 Ile (I) 28 6.7 1.1775 Leu (L) 9.9 Leu (L) 55 13.1 1.3232 Lys (K) 4.93 Lys (K) 23 5.5 1.1156 Met (M) 2.38 Met (M) 6 1.4 0.5882 Phe (F) 3.92 Phe (F) 28 6.7 1.7092 Pro (P) 4.85 Pro (P) 19 4.5 0.9278 Ser (S) 6.64 Ser (S) 25 6 0.9036 Thr (T) 5.55 Thr (T) 13 3.1 0.5586 Trp (W) 1.3 Trp (W) 5 1.2 0.9231 Phe (F) 3.92 Phe (F) 28 6.7 1.7092 Val (V) 6.91 Val (V) 25 6 0.8683 FASTA Files for N-terminals to first zinc finger ZNF292 N-Terminal (to first Zinc Finger) >ZNF292 N-terminal Homo Sapiens MADEEAEQERLSCGEGGCVAELQRLGERLQELELQLRESRVPAVEAATDYCQQLCQTLLE YAEKWKTSEDPLPLLEVYTVAIQSYVKARPYLTSECENVALVLERLALSCVELLLCLPVE LSDKQWEQFQTLVQVAHEKLMENGSCELHFLATLAQETGVWKNPVLCTILSQEPLDKDKV NEFLAFEGPILLDMRIKHLIKTNQLSQATALAKLCSDHPEIGIKGSFKQTYLVCLCTSSP NGKLIEEISEVDCKDALEMICNLESEGDEKSALVLCTAFLSRQLQQGDMYCAWELTLFWS KLQQRVEPSIQVYLERCRQLSLLTKTVYHIFFLIKVINSETEGAGLATCIELCVKALRLE STENTEVKISICKTISCLLPDDLEVKRACQLSEFLIEPTVDAYYAVEMLYNQPDQKYDEE NLPIPNSLRCELLLVLKTQWPFDPEFWDWKTLKRQCLALMGEEASIVSSIDELNDSEVYE KVVDYQEESKETSMNGLSGGVGANSGLLKDIGDEKQKKREIKQLRERGFISARFRNWQAY MQY RLF N-Terminal (to first Zinc Finger) >RLF N-terminal Homo Sapiens MADGKGDAAAVAGAGAEAPAVAGAGDGVETESMVRGHRPVSPAPGASGLRPCLWQLETEL REQEVSEVSSLNYCRSFCQTLLQYASNKNASEHIVYLLEVYRLAIQSFASARPYLTTECE DVLLVLGRLVLSCFELLLSVSESELPCEVWLPFLQSLQESHDALLEFGNNNLQILVHVTK EGVWKNPVLLKILSQQPVETEEVNKLIAQEGPSFLQMRIKHLLKSNCIPQATALSKLCAE SKEISNVSSFQQAYITCLCSMLPNEDAIKEIAKVDCKEVLDIICNLESEGQDNTAFVLCT TYLTQQLQTASVYCSWELTLFWSKLQRRIDPSLDTFLERCRQFGVIAKTQQHLFCLIRVI QTEAQDAGLGVSILLCVRALQLRSSEDEEMKASVCKTIACLLPEDLEVRRACQLTEFLIE PSLDGFNMLEELYLQPDQKFDEENAPVPNSLRCELLLALKAHWPFDPEFWDWKTLKRHCH QLLGQEASDSDDDLSGYEMSINDTDVLESFLSDYDEGKEDKQYRRRDLTDQHKEKRDKKP IGSSERYQRWLQYKFF ZNF654 N-Terminal (to first Zinc Finger) >ZNF654 N-terminal Homo Sapiens MAEEESDQEAERLGEELVAIVESPLGPVGLRAAGDGRGGAGSGNCGGGVGISSRDYCRRF CQVVEDYAGRWQVPLPQLQVLQTALCCFTTASASFPDECEHVQYVLSSLAVSFFELLLFF GRDEFYEEPLKDILGSFQECQNHLRRYGNVNLELVTRIIRDGGPWEDPVLQAVLKAQPAS QEIVNKYLSSENPLFFELRARYLIACERIPEAMALIKSCINHPEISKDLYFHQALFTCLF MSPVEDQLFREHLLKTDCKSGIDIICNAEKEGKTMLALQLCESFLIPQLQNGDMYCIWEL IFIWSKLQLKSNPSKQVFVDQCYQLLRTATNVRVIFPFMKIIKDEVEEEGLQICVEICGC ALQLDLHDDPKTKCLIYKTIAHFLPNDLEILRICALSIFFLERSLEAYRTVEELYKRPDE EYNEGTSSVQNRVRFELLPILKKGLFFDPEFWNFVMIKKNCVALLSDKSAVRFLNESTLE NNAGNLKRTEEQQGLDEGFDSLTDQSTGETDPDDVSGVQPKGHINTKKNLTALSTSKVDH NVPRHR |